User:Bread6/Chemical Biology
| This is the sandbox page where you will draft your initial Wikipedia contribution.
If you're starting a new article, you can develop it here until it's ready to go live. If you're working on improvements to an existing article, copy only one section at a time of the article to this sandbox to work on, and be sure to use an edit summary linking to the article you copied from. Do not copy over the entire article. You can find additional instructions here. Remember to save your work regularly using the "Publish page" button. (It just means 'save'; it will still be in the sandbox.) You can add bold formatting to your additions to differentiate them from existing content. |
Article Draft[edit]
Lead[edit]
Chemical biology is a scientific discipline between the fields of chemistry and biology. The discipline involves the application of chemical techniques, analysis, and often small molecules produced through synthetic chemistry, to the study and manipulation of biological systems.[1] Although often confused with biochemistry, which studies the chemistry of biomolecules and regulation of biochemical pathways within and between cells, chemical biology remains distinct by focusing on the application of chemical tools to address biological questions.[2]

Article body[edit]
History[edit]
Although considered a relatively new scientific field,[2] the term "chemical biology" has been in use since the early 20th century,[3] and has roots in scientific discovery from the early 19th century. The term 'chemical biology' can be traced back to an early appearance in a book published by Alonzo E. Taylor in 1907 titled "On Fermentation",[4] and was subsequently used in John B. Leathes' 1930 article titled "The Harveian Oration on The Birth of Chemical Biology".[5] However, it is unclear when the term was first used.[6]
Friedrich Wöhler's 1828 synthesis of urea is an early example of the application of synthetic chemistry to advance biology.[7] It showed that biological compounds could be synthesized with inorganic starting materials and weakened the previous notion of vitalism, or that a 'living' source was required to produce organic compounds.[8][9] Wöhler's work is often considered to be instrumental in the development of organic chemistry and natural product synthesis, both of which play a large part in modern chemical biology.[10]
Friedrich Miescher's work during the late 19th century investigating the cellular contents of human leukocytes led to the discovery of 'nuclein', which would later be renamed DNA.[7] After isolating the nuclein from the nucleus of leukocytes through protease digestion, Miescher used chemical techniques such as elemental analysis and solubility tests to determine the composition of nuclein.[11] This work would lay the foundations for Watson and Crick's discovery of the double-helix structure of DNA.[11][12]
The rising interest into chemical biology has led to the creation of multiple journals dedicated to the field. Nature Chemical Biology, created in 2005,[13] and ACS Chemical Biology, created in 2006,[14] are two of the most well-known journals in this field, with impact factors of 14.8[15] and 4.0[16] respectively.
Nobel laureates in chemical biology[edit]
| Laureate | Year | Discipline | Contribution |
|---|---|---|---|
| Paul Berg | 1980 | Chemistry | Recombinant DNA[17] |
| Walter Gilbert | 1980 | Chemistry | Genome sequencing[17] |
| Kary Mullis | 1993 | Chemistry | Polymerase chain reaction[18] |
| Michael Smith | 1993 | Chemistry | Site-directed mutagenesis[18] |
| Venkatraman Ramakrishnan | 2009 | Chemistry | Elucidation of ribosome structure and function[19] |
| Robert J. Lefkowitz | 2012 | Chemistry | G-protein-coupled receptors[20] |
| Frances H. Arnold | 2018 | Chemistry | Enzyme development through directed evolution[21] |
| Emmanuelle Charpentier | 2020 | Chemistry | CRISPR/Cas9 genetic scissors[22] |
| Barry Sharpless | 2022 | Chemistry | Click chemistry[23] |
| Carolyn Bertozzi | 2022 | Chemistry | Applications of click chemistry in living organisms[23] |
Education in Chemical Biology[edit]
Undergraduate Education[edit]
Despite an increase in biological research within chemistry departments, attempts at integrating chemical biology into undergraduate curricula are lacking.[24] For example, although the American Chemical Society (ACS) requires for foundational courses in a Chemistry Bachelor's degree to include biochemistry, no other biology-related chemistry course is required.[25]
Although a chemical biology course is often not required for an undergraduate degree in Chemistry, many universities now provide introductory chemical biology courses for their undergraduate students. The University of British Columbia, for example, offers a fourth-year course in synthetic chemical biology.[26]
Research Areas[edit]
Glycobiology[edit]

Glycobiology is the study of the structure and function of carbohydrates.[27] While DNA, RNA and proteins are encoded at the genetic level, carbohydrates are not encoded directly from the genome, and thus require different tools for their study.[28] By applying chemical principles to glycobiology, novel methods for analyzing and synthesizing carbohydrates can be developed.[29] For example, cells can be supplied with synthetic variants of natural sugars to probe their function. Carolyn Bertozzi's research group has developed methods for site-specifically reacting molecules at the surface of cells via synthetic sugars.[30]
Combinatorial chemistry[edit]
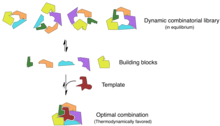
Combinatorial chemistry involves simultaneously synthesizing a large number of related compounds for high-throughput analysis.[31] Chemical biologists are able to use principles from combinatorial chemistry in synthesizing active drug compounds and maximizing screening efficiency.[32] Similarly, these principles can be used in areas of agriculture and food research, specifically in the syntheses of unnatural products and in generating novel enzyme inhibitors.[33]
Peptide synthesis[edit]

Chemical synthesis of proteins is a valuable tool in chemical biology as it allows for the introduction of non-natural amino acids as well as residue specific incorporation of "posttranslational modifications" such as phosphorylation, glycosylation, acetylation, and even ubiquitination.[34] These properties are valuable for chemical biologists as non-natural amino acids can be used to probe and alter the functionality of proteins, while post-translational modifications are widely known to regulate the structure and activity of proteins.[35] Although strictly biological techniques have been developed to achieve these ends, the chemical synthesis of peptides often has a lower technical and practical barrier to obtaining small amounts of the desired protein.[36]
To make protein-sized polypeptide chains with the small peptide fragments made by synthesis, chemical biologists can use the process of native chemical ligation.[37] Native chemical ligation involves the coupling of a C-terminal thioester and an N-terminal cysteine residue, ultimately resulting in formation of a "native" amide bond.[38] Other strategies that have been used for the ligation of peptide fragments using the acyl transfer chemistry first introduced with native chemical ligation include expressed protein ligation,[39] sulfurization/desulfurization techniques,[40] and use of removable thiol auxiliaries.[41]
Bioorthogonal reactions[edit]
Successful labeling of a molecule of interest requires specific functionalization of that molecule to react chemospecifically with an optical probe. For a labeling experiment to be considered robust, that functionalization must minimally perturb the system. Unfortunately, these requirements are often hard to meet. Many of the reactions normally available to organic chemists in the laboratory are unavailable in living systems.[42] Water- and redox- sensitive reactions would not proceed, reagents prone to nucleophilic attack would offer no chemospecificity, and any reactions with large kinetic barriers would not find enough energy in the relatively low-heat environment of a living cell.[43] Thus, chemists have recently developed a panel of bioorthogonal chemistry that proceed chemospecifically, despite the milieu of distracting reactive materials in vivo.
The coupling of a probe to a molecule of interest must occur within a reasonably short time frame[44]; therefore, the kinetics of the coupling reaction should be highly favorable. Click chemistry is well suited to fill this niche, since click reactions are rapid, spontaneous, selective, and high-yielding. Unfortunately, the most famous "click reaction," a [3+2] cycloaddition between an azide and an acyclic alkyne, is copper-catalyzed, posing a serious problem for use in vivo due to copper's toxicity. To bypass the necessity for a catalyst, Carolyn R. Bertozzi's lab introduced inherent strain into the alkyne species by using a cyclic alkyne. In particular, cyclooctyne reacts with azido-molecules with distinctive vigor.
Directed evolution[edit]
A primary goal of protein engineering is the design of novel peptides or proteins with a desired structure and chemical activity.[45] Because our knowledge of the relationship between primary sequence, structure, and function of proteins is limited, rational design of new proteins with engineered activities is extremely challenging.[46] In directed evolution, repeated cycles of genetic diversification followed by a screening or selection process, can be used to mimic natural selection in the laboratory to design new proteins with a desired activity.[47]
Several methods exist for creating large libraries of sequence variants. Among the most widely used are subjecting DNA to UV radiation or chemical mutagens, error-prone PCR, degenerate codons, or recombination.[48][49] Once a large library of variants is created, selection or screening techniques are used to find mutants with a desired attribute. Common selection/screening techniques include FACS,[50] mRNA display,[51] phage display, and in vitro compartmentalization.[52] Once useful variants are found, their DNA sequence is amplified and subjected to further rounds of diversification and selection.
The development of directed evolution methods was honored in 2018 with the awarding of the Nobel Prize in Chemistry to Frances Arnold for evolution of enzymes, and George Smith and Gregory Winter for phage display.[53]
References[edit]
- ^ Schreiber, Stuart L. (2005-07). "Small molecules: the missing link in the central dogma". Nature Chemical Biology. 1 (2): 64–66. doi:10.1038/nchembio0705-64. ISSN 1552-4469.
{{cite journal}}: Check date values in:|date=(help) - ^ a b Miller, Andrew; Tanner, Julian (2008). Essentials of Chemical Biology: Structure and Dynamics of Biological Macromolecules. England: John Wiley & Sons, Ltd. pp. vi–x. ISBN 978-0-470-84530-1.
- ^ Hricovini, Milos; Jampilek, Josef (2023-02-16). "Chemistry towards Biology". International Journal of Molecular Sciences. 24 (4): 3998. doi:10.3390/ijms24043998. ISSN 1422-0067. PMC 9960482. PMID 36835407.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: unflagged free DOI (link) - ^ Taylor, Alonzo Englebert (1907). On fermentation. 8. Vol. 1. University Press.
- ^ Leathes, J. B. (1930-10-25). "The Harveian Oration on THE BIRTH OF CHEMICAL BIOLOGY". BMJ. 2 (3642): 671–676. doi:10.1136/bmj.2.3642.671. ISSN 0959-8138. PMC 2451377. PMID 20775787.
{{cite journal}}: CS1 maint: PMC format (link) - ^ Hricovini, Milos; Jampilek, Josef (2023-02-16). "Chemistry towards Biology". International Journal of Molecular Sciences. 24 (4): 3998. doi:10.3390/ijms24043998. ISSN 1422-0067. PMC 9960482. PMID 36835407.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: unflagged free DOI (link) - ^ a b Morrison, Kim L.; Weiss, Gregory A. (2006-01). "The origins of chemical biology". Nature Chemical Biology. 2 (1): 3–6. doi:10.1038/nchembio0106-3. ISSN 1552-4469.
{{cite journal}}: Check date values in:|date=(help) - ^ Ramberg, Peter J. (2000-11). "The Death of Vitalism and The Birth of Organic Chemistry: Wohler's Urea Synthesis and the Disciplinary Identity of Organic Chemistry". Ambix. 47 (3): 170–195. doi:10.1179/amb.2000.47.3.170. ISSN 0002-6980.
{{cite journal}}: Check date values in:|date=(help) - ^ www.karger.com. doi:10.1159/000013463 https://www.karger.com/Article/FullText/13463. Retrieved 2023-11-05.
{{cite web}}: Missing or empty|title=(help) - ^ Hong, Jiyong (2014-08-11). "Natural Product Synthesis at the Interface of Chemistry and Biology". Chemistry – A European Journal. 20 (33): 10204–10212. doi:10.1002/chem.201402804. ISSN 0947-6539. PMC 4167019. PMID 25043880.
{{cite journal}}: CS1 maint: PMC format (link) - ^ a b Dahm, Ralf (2008-01-01). "Discovering DNA: Friedrich Miescher and the early years of nucleic acid research". Human Genetics. 122 (6): 565–581. doi:10.1007/s00439-007-0433-0. ISSN 1432-1203.
- ^ Dahm, Ralf (2005-02-15). "Friedrich Miescher and the discovery of DNA". Developmental Biology. 278 (2): 274–288. doi:10.1016/j.ydbio.2004.11.028. ISSN 0012-1606.
- ^ "LC Catalog - Item Information (Full Record)". catalog.loc.gov. Retrieved 2023-11-06.
- ^ "LC Catalog - Item Information (Full Record)". catalog.loc.gov. Retrieved 2023-11-06.
- ^ "Journal Metrics | Nature Chemical Biology". www.nature.com. Retrieved 2023-11-06.
- ^ "About the Journal". ACS Publications. Retrieved November 5, 2023.
- ^ a b "The Nobel Prize in Chemistry 1980". NobelPrize.org. Retrieved 2023-11-06.
- ^ a b "The Nobel Prize in Chemistry 1993". NobelPrize.org. Retrieved 2023-11-06.
- ^ "The Nobel Prize in Chemistry 2009". NobelPrize.org. Retrieved 2023-11-06.
- ^ "The Nobel Prize in Chemistry 2012". NobelPrize.org. Retrieved 2023-11-06.
- ^ "The Nobel Prize in Chemistry 2018". NobelPrize.org. Retrieved 2023-11-06.
- ^ "The Nobel Prize in Chemistry 2020". NobelPrize.org. Retrieved 2023-11-06.
- ^ a b "The Nobel Prize in Chemistry 2022". NobelPrize.org. Retrieved 2023-11-06.
- ^ Begley, Tadhg P. (2005-10). "Chemical biology: an educational challenge for chemistry departments". Nature Chemical Biology. 1 (5): 236–238. doi:10.1038/nchembio1005-236. ISSN 1552-4469.
{{cite journal}}: Check date values in:|date=(help) - ^ "Coursework". American Chemical Society. Retrieved 2023-12-02.
- ^ "Chemistry 461: Synthetic Chemical Biology | UBC Chemistry". www.chem.ubc.ca. Retrieved 2023-12-02.
- ^ Dwek, Raymond A. (1996-01-01). "Glycobiology: Toward Understanding the Function of Sugars". Chemical Reviews. 96 (2): 683–720. doi:10.1021/cr940283b. ISSN 0009-2665.
- ^ Springer, Stevan A.; Gagneux, Pascal (2016-03). "Glycomics: revealing the dynamic ecology and evolution of sugar molecules". Journal of Proteomics. 135: 90–100. doi:10.1016/j.jprot.2015.11.022. PMC 4762723. PMID 26626628.
{{cite journal}}: Check date values in:|date=(help)CS1 maint: PMC format (link) - ^ Wu, Chung-Yi; Wong, Chi-Huey (2011). "Chemistry and glycobiology". Chemical Communications. 47 (22): 6201. doi:10.1039/c0cc04359a. ISSN 1359-7345.
- ^ "Bertozzi Group". Bertozzi Group. Retrieved 2023-12-04.
- ^ Liu, Ruiwu; Li, Xiaocen; Lam, Kit S (2017-06). "Combinatorial chemistry in drug discovery". Current Opinion in Chemical Biology. 38: 117–126. doi:10.1016/j.cbpa.2017.03.017. PMC 5645069. PMID 28494316.
{{cite journal}}: Check date values in:|date=(help)CS1 maint: PMC format (link) - ^ Kennedy, J. Phillip; Williams, Lyndsey; Bridges, Thomas M.; Daniels, R. Nathan; Weaver, David; Lindsley, Craig W. (2008-05-01). "Application of Combinatorial Chemistry Science on Modern Drug Discovery". Journal of Combinatorial Chemistry. 10 (3): 345–354. doi:10.1021/cc700187t. ISSN 1520-4766.
- ^ Wong, Dominic; Robertson, George (2004-12-01). "Applying Combinatorial Chemistry and Biology to Food Research". Journal of Agricultural and Food Chemistry. 52 (24): 7187–7198. doi:10.1021/jf040140i. ISSN 0021-8561.
- ^ Ramazi, Shahin; Zahiri, Javad (2021-04-07). "Post-translational modifications in proteins: resources, tools and prediction methods". Database. 2021. doi:10.1093/database/baab012. ISSN 1758-0463. PMC 8040245. PMID 33826699.
{{cite journal}}: CS1 maint: PMC format (link) - ^ Adhikari, Anup; Bhattarai, Bibek Raj; Aryal, Ashika; Thapa, Niru; Kc, Puja; Adhikari, Ashma; Maharjan, Sushila; Chanda, Prem B.; Regmi, Bishnu P.; Parajuli, Niranjan (2021). "Reprogramming natural proteins using unnatural amino acids". RSC Advances. 11 (60): 38126–38145. doi:10.1039/D1RA07028B. ISSN 2046-2069. PMC 9044140. PMID 35498070.
{{cite journal}}: CS1 maint: PMC format (link) - ^ Chandrudu, Saranya; Simerska, Pavla; Toth, Istvan (2013-04-12). "Chemical Methods for Peptide and Protein Production". Molecules. 18 (4): 4373–4388. doi:10.3390/molecules18044373. ISSN 1420-3049. PMC 6270108. PMID 23584057.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: unflagged free DOI (link) - ^ Cistrone, Philip A.; Bird, Michael J.; Flood, Dillon T.; Silvestri, Anthony P.; Hintzen, Jordi C. J.; Thompson, Darren A.; Dawson, Philip E. (2019-03). "Native Chemical Ligation of Peptides and Proteins". Current Protocols in Chemical Biology. 11 (1). doi:10.1002/cpch.61. ISSN 2160-4762. PMC 6384150. PMID 30645048.
{{cite journal}}: Check date values in:|date=(help)CS1 maint: PMC format (link) - ^ Hermanson, Greg T. (2013-01-01), Hermanson, Greg T. (ed.), "Chapter 3 - The Reactions of Bioconjugation", Bioconjugate Techniques (Third Edition), Boston: Academic Press, pp. 229–258, doi:10.1016/b978-0-12-382239-0.00003-0, ISBN 978-0-12-382239-0, retrieved 2023-12-04
- ^ Muir, Tom W.; Sondhi, Dolan; Cole, Philip A. (1998-06-09). "Expressed protein ligation: A general method for protein engineering". Proceedings of the National Academy of Sciences. 95 (12): 6705–6710. doi:10.1073/pnas.95.12.6705. ISSN 0027-8424. PMC 22605. PMID 9618476.
{{cite journal}}: CS1 maint: PMC format (link) - ^ Jin, Kang; Li, Tianlu; Chow, Hoi Yee; Liu, Han; Li, Xuechen (2017-11-13). "P−B Desulfurization: An Enabling Method for Protein Chemical Synthesis and Site‐Specific Deuteration". Angewandte Chemie International Edition. 56 (46): 14607–14611. doi:10.1002/anie.201709097. ISSN 1433-7851.
- ^ Nilsson, Bradley L.; Soellner, Matthew B.; Raines, Ronald T. (2005-06-01). "Chemical Synthesis of Proteins". Annual Review of Biophysics and Biomolecular Structure. 34 (1): 91–118. doi:10.1146/annurev.biophys.34.040204.144700. ISSN 1056-8700. PMC 2845543. PMID 15869385.
{{cite journal}}: CS1 maint: PMC format (link) - ^ Jonsson, Amanda L.; Roberts, Mark A.J.; Kiappes, J.L.; Scott, Kathryn A. (2017-10-31). "Essential chemistry for biochemists". Essays in Biochemistry. 61 (4): 401–427. doi:10.1042/EBC20160094. ISSN 0071-1365. PMC 5869253. PMID 28951470.
{{cite journal}}: no-break space character in|first2=at position 5 (help); no-break space character in|first4=at position 8 (help); no-break space character in|first=at position 7 (help)CS1 maint: PMC format (link) - ^ Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Raff, Martin; Roberts, Keith; Walter, Peter (2002), "Catalysis and the Use of Energy by Cells", Molecular Biology of the Cell. 4th edition, Garland Science, retrieved 2023-12-06
- ^ Chen, Kai; Chen, Xiaoyuan. "Design and Development of Molecular Imaging Probes". Current Topics in Medicinal Chemistry. 10 (12): 1227–1236. doi:10.2174/156802610791384225. PMC 3632640. PMID 20388106.
{{cite journal}}: CS1 maint: PMC format (link) - ^ Dhanjal, Jaspreet Kaur; Malik, Vidhi; Radhakrishnan, Navaneethan; Sigar, Moolchand; Kumari, Anjani; Sundar, Durai (2019-01-01), Ranganathan, Shoba; Gribskov, Michael; Nakai, Kenta; Schönbach, Christian (eds.), "Computational Protein Engineering Approaches for Effective Design of New Molecules", Encyclopedia of Bioinformatics and Computational Biology, Oxford: Academic Press, pp. 631–643, doi:10.1016/b978-0-12-809633-8.20150-7, ISBN 978-0-12-811432-2, retrieved 2023-12-06
- ^ Kuhlman, Brian; Bradley, Philip (2019-11). "Advances in protein structure prediction and design". Nature Reviews Molecular Cell Biology. 20 (11): 681–697. doi:10.1038/s41580-019-0163-x. ISSN 1471-0080. PMC 7032036. PMID 31417196.
{{cite journal}}: Check date values in:|date=(help)CS1 maint: PMC format (link) - ^ Jäckel C, Kast P, Hilvert D (2008). "Protein design by directed evolution". Annual Review of Biophysics. 37: 153–173. doi:10.1146/annurev.biophys.37.032807.125832. PMID 18573077.
- ^ Taylor SV, Walter KU, Kast P, Hilvert D (September 2001). "Searching sequence space for protein catalysts". Proceedings of the National Academy of Sciences of the United States of America. 98 (19): 10596–10601. Bibcode:2001PNAS...9810596T. doi:10.1073/pnas.191159298. PMC 58511. PMID 11535813.
- ^ Bittker JA, Le BV, Liu JM, Liu DR (May 2004). "Directed evolution of protein enzymes using nonhomologous random recombination". Proceedings of the National Academy of Sciences of the United States of America. 101 (18): 7011–7016. Bibcode:2004PNAS..101.7011B. doi:10.1073/pnas.0402202101. PMC 406457. PMID 15118093.
- ^ Aharoni A, Griffiths AD, Tawfik DS (April 2005). "High-throughput screens and selections of enzyme-encoding genes". Current Opinion in Chemical Biology. 9 (2): 210–216. doi:10.1016/j.cbpa.2005.02.002. PMID 15811807.
- ^ Wilson DS, Keefe AD, Szostak JW (March 2001). "The use of mRNA display to select high-affinity protein-binding peptides". Proceedings of the National Academy of Sciences of the United States of America. 98 (7): 3750–3755. Bibcode:2001PNAS...98.3750W. doi:10.1073/pnas.061028198. PMC 31124. PMID 11274392.
- ^ Tawfik DS, Griffiths AD (July 1998). "Man-made cell-like compartments for molecular evolution". Nature Biotechnology. 16 (7): 652–656. doi:10.1038/nbt0798-652. PMID 9661199. S2CID 25527137.
- ^ "The Nobel Prize in Chemistry 2018". NobelPrize.org. Retrieved 2018-10-03.
