User:Wjwalker17/sandbox
| xanthine oxidase/dehydrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Crystallographic structure (monomer) of bovine xanthine oxidase.[1] The bounded FAD (red), FeS-cluster (orange), the molybdopterin cofactor with molybdenum (yellow) and salicylate (blue) are indicated. | |||||||||
| Identifiers | |||||||||
| EC no. | 1.17.3.2 | ||||||||
| CAS no. | 9002-17-9 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| xanthine oxidase/dehydrogenase | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Symbol | XDH | ||||||
| NCBI gene | 7498 | ||||||
| HGNC | 12805 | ||||||
| OMIM | 607633 | ||||||
| PDB | 1FIQ | ||||||
| RefSeq | NM_000379 | ||||||
| UniProt | P47989 | ||||||
| Other data | |||||||
| EC number | 1.17.3.2 | ||||||
| Locus | Chr. 2 p23.1 | ||||||
| |||||||
Xanthine oxidase (XO, sometimes 'XAO') is a form of xanthine oxidoreductase, a type of enzyme that generates reactive oxygen species.[2] These enzymes catalyze the oxidation of hypoxanthine to xanthine and can further catalyze the oxidation of xanthine to uric acid. These enzymes play an important role in the catabolism of purines in some species, including humans.[3][4]
Xanthine oxidase is defined as an enzyme activity (EC 1.17.3.2).[5] The same protein, which in humans has the HGNC approved gene symbol XDH, can also have xanthine dehydrogenase activity (EC 1.17.1.4).[6] Most of the protein in the liver exists in a form with xanthine dehydrogenase activity, but it can be converted to xanthine oxidase by reversible sulfhydryl oxidation or by irreversible proteolytic modification.[7][8]
Reaction[edit]
The following chemical reactions are catalyzed by xanthine oxidase:
- hypoxanthine + H2O + O2 xanthine + H2O2
- xanthine + H2O + O2 uric acid + H2O2
- Xanthine oxidase can also act on certain other purines, pterins, and aldehydes. For example, it efficiently converts 1-methylxanthine (a metabolite of caffeine) to 1-methyluric acid, but has little activity on 3-methylxanthine.[9]
- Under some circumstances it can produce superoxide ion RH + H2O + 2 O2 ROH + 2 O2− + 2 H+.[6]
-
hypoxanthine (one oxygen atom)
-
xanthine (two oxygens)
-
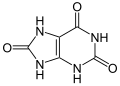
uric acid (three oxygens)
Protein structure[edit]
The protein is large, having a molecular weight of 270 kDa, and has 2 flavin molecules (bound as FAD), 2 molybdenum atoms, and 8 iron atoms bound per enzymatic unit. The molybdenum atoms are contained as molybdopterin cofactors and are the active sites of the enzyme. The iron atoms are part of [2Fe-2S] ferredoxin iron-sulfur clusters and participate in electron transfer reactions.
Catalytic mechanism[edit]
The active site of XO is composed of a molybdopterin unit with the molybdenum atom also coordinated by terminal oxygen (oxo), sulfur atoms and a terminal hydroxide.[10] In the reaction with xanthine to form uric acid, an oxygen atom is transferred from molybdenum to xanthine, whereby several intermediates are assumed to be involved.[11] The reformation of the active molybdenum center occurs by the addition of water. Like other known molybdenum-containing oxidoreductases, the oxygen atom introduced to the substrate by XO originates from water rather than from dioxygen (O2).
Clinical significance[edit]
Xanthine oxidase is a superoxide-producing enzyme found normally in serum and the lungs, and its activity is increased during influenza A infection.[12]
During severe liver damage, xanthine oxidase is released into the blood, so a blood assay for XO is a way to determine if liver damage has happened.[citation needed]
As well, because xanthine oxidase is a metabolic pathway for uric acid formation, the xanthine oxidase inhibitor allopurinol is used in the treatment of gout. Since xanthine oxidase is involved in the metabolism of 6-mercaptopurine, caution should be taken before administering allopurinol and 6-mercaptopurine, or its prodrug azathioprine, in conjunction.
Xanthinuria is a rare genetic disorder where the lack of xanthine oxidase leads to high concentration of xanthine in blood and can cause health problems such as renal failure. There is no specific treatment, sufferers are advised by doctors to avoid foods high in purine and to maintain a high fluid intake. Type I xanthinuria has been traced directly to mutations of the XDH gene which mediates xanthine oxidase activity. Type II xanthinuria may result from a failure of the mechanism which inserts sulfur into the active sites of xanthine oxidase and aldehyde oxidase, a related enzyme with some overlapping activities (such as conversion of allopurinol to oxypurinol).[13]
Inhibition of xanthine oxidase has been proposed as a mechanism for improving cardiovascular health.[14]
Both xanthine oxidase and xanthine oxidoreductase are also present in corneal epithelium and endothelium and may be involved in oxidative eye injury.[15]
Inhibitors[edit]
Inhibitors of XO include allopurinol,[16] oxypurinol,[17] and phytic acid.[18] It has also been found to be inhibited by flavonoids[19], including those found in Bougainvillea spectabilis Willd (Nyctaginaceae) leaves (with an IC50 of 7.23 μM), typically used in folk medicine[20].
See also[edit]
References[edit]
- ^ PDB: 1FIQ; Enroth C, Eger BT, Okamoto K, Nishino T, Nishino T, Pai EF (September 2000). "Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion". Proc. Natl. Acad. Sci. U.S.A. 97 (20): 10723–8. doi:10.1073/pnas.97.20.10723. PMC 27090. PMID 11005854.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ardan T, Kovaceva J, Cejková J (2004). "Comparative histochemical and immunohistochemical study on xanthine oxidoreductase/xanthine oxidase in mammalian corneal epithelium". Acta Histochem. 106 (1): 69–75. doi:10.1016/j.acthis.2003.08.001. PMID 15032331.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hille R (2005). "Molybdenum-containing hydroxylases". Arch. Biochem. Biophys. 433 (1): 107–16. doi:10.1016/j.abb.2004.08.012. PMID 15581570.
- ^ Harrison R (2002). "Structure and function of xanthine oxidoreductase: where are we now?". Free Radic. Biol. Med. 33 (6): 774–97. doi:10.1016/S0891-5849(02)00956-5. PMID 12208366.
- ^ "KEGG record for EC 1.17.3.2".
- ^ a b "KEGG record for EC 1.17.1.4".
- ^ "Entrez Gene: XDH xanthine dehydrogenase".
- ^ "*607633 XANTHINE DEHYDROGENASE; XDH".
- ^ D. J. Birkett; et al. (1997). "1-Methylxanthine derived from caffeine as a pharmacodynamic probe of oxypurinol effect". Br J Clin Pharmacol. 43 (2): 197–200. doi:10.1046/j.1365-2125.1997.53711.x. PMC 2042732. PMID 9131954.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ Hille R. (2006). "Structure and Function of Xanthine Oxidoreductase". European Journal of Inorganic Chemistry. 2006 (10): 1905–2095. doi:10.1002/ejic.200600087.
- ^ Metz S, Thiel W. (2009). "A Combined QM/MM Study on the Reductive Half-Reaction of Xanthine Oxidase: Substrate Orientation and Mechanism". Journal of the American Chemical Society. 131 (41): 14885–158902. doi:10.1021/ja9045394. PMID 19788181.
- ^ http://www.colorado.edu/intphys/iphy3700/vitCHemila92.pdf
- ^ "OMIM: Xanthinuria type II".
- ^ Dawson J, Walters M (October 2006). "Uric acid and xanthine oxidase: future therapeutic targets in the prevention of cardiovascular disease?". British Journal of Clinical Pharmacology. 62 (6): 633–644. doi:10.1111/j.1365-2125.2006.02785.x. PMC 1885190. PMID 21894646.
- ^ Cejková J, Ardan T, Filipec M, Midelfart A (2002). "Xanthine oxidoreductase and xanthine oxidase in human cornea". Histol. Histopathol. 17 (3): 755–60. PMID 12168784.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Pacher P, Nivorozhkin A, Szabó C (March 2006). "Therapeutic effects of xanthine oxidase inhibitors: renaissance half a century after the discovery of allopurinol". Pharmacol. Rev. 58 (1): 87–114. doi:10.1124/pr.58.1.6. PMC 2233605. PMID 16507884.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Spector T (January 1988). "Oxypurinol as an inhibitor of xanthine oxidase-catalyzed production of superoxide radical". Biochem. Pharmacol. 37 (2): 349–52. doi:10.1016/0006-2952(88)90739-3. PMID 2829916.
- ^ Muraoka S, Miura T (February 2004). "Inhibition of xanthine oxidase by phytic acid and its antioxidative action". Life Sci. 74 (13): 1691–700. doi:10.1016/j.lfs.2003.09.040. PMID 14738912.
- ^ Cos, Paul; Ying, Li; Calomme, Mario; Hu, Jia P.; Cimanga, Kanyanga; Van Poel, Bart; Pieters, Luc; Vlietinck, Arnold J.; Berghe, Dirk Vanden (January 1998). "Structure−Activity Relationship and Classification of Flavonoids as Inhibitors of Xanthine Oxidase and Superoxide Scavengers". Journal of Natural Products. 61 (1): 71–76. doi:10.1021/np970237h. PMID 9461655.
- ^ Chang, W. S.; Lee, Y. J.; Lu, F. J.; Chiang, H. C. (1993 Nov-Dec). "Inhibitory effects of flavonoids on xanthine oxidase". Anticancer Research. 13 (6A): 2165–70. PMID 8297130.
{{cite journal}}: Check date values in:|date=(help)
External links[edit]
- Xanthine+Oxidase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
Category:EC 1.17.3 Category:Metalloproteins Category:Molybdenum enzymes Category:Iron-sulfur enzymes