User:Solarys-fr/Gut flora
| This is not a Wikipedia article: It is an individual user's work-in-progress page, and may be incomplete and/or unreliable. For guidance on developing this draft, see Wikipedia:So you made a userspace draft. Find sources: Google (books · news · scholar · free images · WP refs) · FENS · JSTOR · TWL |

Gut flora or gut microbiota is a complex community of microorganisms that live in the digestive tract of animals and humans.
In this context, gut covers a wide meaning, because it refers not only to intestine but also to the rest of the digestive tract, including the mouth. Microflora is synonymous with microbiota. A related term, sometimes used interchangeably with gut microbiota, is gut microbiome, which refers to the aggregate of all of the genomes of gut microbiota. The genes these microorganisms encode form the metagenome,[1] known as the "second genome".[2][3] This collective genome is 150 times larger than the human genome.[4]
Microorganisms live on and inside the cavities of the human body[5] but the highest density and diversity of microorganisms is located in the human intestinal tract or gut,[5] which harbors 10*14 microorganisms per gram of intestinal content. The human gut contains hundreds of bacterial species and many times more genes than are found in the human genome.[6]
The metabolic activities performed by these bacteria resemble those of an organ, leading researchers to consider gut microbiota as a "forgotten" or "hidden" organ.[7][8][5]
Research into the association between the gut microbiota and health has increased in recent years and continues to expand.[9][5]
The gut microbiota interacts with the host: locally with the mucosa and the intestinal barrier as well as the immune system and, thus, with the whole body. The gut microbiota may influence the development of several diseases[1] and plays a central role in human health.[9][5][6]
While an individual's inherited genes are fixed, one's second genome is changing constantly according to age, as well as diet and environmental factors.[3]
Description[edit]
Until recently, researchers believed the human body was composed only of human DNA. In reality, the genetic capital includes the billions of strands of DNA carried by microorganisms. More and more researchers are talking about the human body as a superorganism, consisting of the human host plus its microbiota.[10]
The human body harbors an ecosystem of bacteria. Distinct microbial communities[7] colonize all surfaces of human body. As in other mammals, trillions of bacteria live on and in the whole body: on the skin, in the mouth, in the vagina, in the lungs and in the intestines[3][11][12][13][14][15] (but not other sites such as the urinary tract or the meninges).
An effort to better describe the microflora of the gut and other body locations was initiated in 2008: the Human microbiome project[16]. In 2009, scientists from INRA (France) highlighted the existence of a small number of species shared by all individuals that constitute the human intestinal microbiota core of functional genes.[17][18][19][20]
Bacterial communities at a given body site across individuals resemble each other more than those at different body sites, i.e., oral bacterial communities across individuals are more similar than the bacterial communities of the skin and the mouth of the same individual. The composition of the microbiota remains relatively stable within healthy adult individuals over time.[7]
Bacteria interact both with one another and with the host. They have individual needs and are dependent on the host for their existence.[21] The human body and its flora therefore have a mutualistic relationship.[22]
The vast majority of the bacteria in the gastrointestinal tract reside in the large intestine[21][6][23]. The small intestine contains a lower abundance and different composition of bacteria, with a more dynamic variation than those in the colon.[24]
Gut microbiota is composed of: bacteria, viruses, eukaryotes (protozoa, fungi)[7][25] and archaea[1].
Over 99% of the bacteria in the gut are anaerobes.[18][19][20][26][18]
The composition of microbiota depends on several factors such as the host's diet, colonization history (birth delivery), lifespan, immune status and medications.[27]
Composition can also vary according to the sex and the geographical origin of the individual.[2][6]
The gut microbiota influences human physiology, digestion, nutrient and drug metabolism, the immune system,[6][23] detoxification, and vitamin production, and may also play a role in the prevention of diseases.[2] The gut microbiota also influences mind and behavior.
Changes to the gut microbiota can have major consequences, both beneficial and harmful to human health.
Disruption of the gut microbiota, called "dysbiosis", often consists of a loss of diversity and increases or decreases in specific microbes. It is a change of diversity and richness. Dysbiosis is linked to various diseases that can affect the body and brain.[3][11][7] Dysbiosis has been linked with gastrointestinal conditions such as obesity, diabetes, and chronic inflammatory diseases such as inflammatory bowel disease (IBD), ulcerative colitis and Crohn’s disease[6][28] as well as cancer, cardiovascular disease and some mental health disorders.[5][6]
Gut microbiota composition can serve as a biomarker for nutritional habits as well as for disease risks. A study published in 2016 has revealed that gut bacteria are critically important, not only for indicating the risk of disease but also for mediating the link between diet and disease, thus assigning the gut microbes a place in the causal chain of disease development.[29]
Bacterial composition[edit]
The total number of bacterial species in the human body has been estimated at over 1,000.[25][19] The human gut can harbor around 200 species of bacteria and approximately 160 species live in the large intestine.[24][30][31][32]
In the human intestinal tract, most bacterial species belong to six dominant phyla: Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria[33], Verrucomicrobia and Fusobacteria[21].
Archaea constitute another large class of gut microorganisms, which are important in the metabolism of the bacterial products of fermentation. Methanogenic archaea called Methanobrevibacter smithii also make part of gut flora.[25][2]
Three phyla dominate the gut flora, seemingly dependent on the diet: Bacteroidetes, Firmicutes, and to a lesser extent Actinobacteria.[23][7][1] One phyla is dominant for each individual.
About 20 core genera are observed[34], with most bacteria belonging to the genera Bacteroides, Clostridium, Faecalibacterium,[18][20][26] Eubacterium, Ruminococcus, Peptococcus, Peptostreptococcus, and Bifidobacterium.[18][20] Species from the genus Bacteroides alone constitute about 30% of all bacteria in the gut.[19]
Fungal genera that have been detected in the gut include Candida, Saccharomyces, Aspergillus, Penicillium, Rhodotorula, Trametes, Pleospora, Sclerotinia, Bullera, and Galactomyces, among others.[35][36] Rhodotorula is most frequently found in individuals with inflammatory bowel disease while Candida is most frequently found in individuals with hepatitis B, cirrhosis and chronic hepatitis B.[35]
Enterotypes[edit]
Populations of gut microorganisms vary widely among different individuals but stay fairly constant within an individual over time, even though some alterations may occur with age and lifestyle factors.[18][31]
The gut microbiota of most individuals can be categorized into one of three variants or “enterotypes”, based on the dominant genera: Bacteroides, Prevotella or Firmicutes.[7][1]
An enterotype is a classification of the gut ecosystem that is not dictated by age, gender, body weight, or country of residence.[37] There are indications that long-term diet influences enterotype.[38]
Prevotella and Firmicutes enterotypes are associated with plant-rich diets,[39][23] whereas the Bacteroides enterotype is often associated with a high-fat, high-protein (meat-based) diet, typical in North America.[23][23] On this diet, Bacteroidetes usually make up more than 55% of the gut microbiota, with the percentage sometimes reaching 80%.[23]
The three human enterotypes were discovered in 2011.[37][40] However, the concept of enterotypes is still under scientific discussion.
Flora composition[edit]
Anatomy[edit]
Stomach flora[edit]
Due to the high acidity of the stomach, most microorganisms cannot survive. The main bacterial inhabitants of the stomach include: Streptococcus, Staphylococcus, Lactobacillus, Peptostreptococcus, and types of yeast.[41]: 720 Helicobacter pylori is a Gram-negative spiral organism that establishes on gastric mucosa causing chronic gastritis and peptic ulcer disease and is a carcinogen for gastric cancer.[41]: 904
Intestinal flora[edit]
| Bacteria commonly found in the human colon[42] | |
| Bacterium | Incidence (%) |
|---|---|
| Bacteroides fragilis | 100 |
| Bacteroides melaninogenicus | 100 |
| Bacteroides oralis | 100 |
| Enterococcus faecalis | 100 |
| Escherichia coli | 100 |
| Enterobacter sp. | 40–80 |
| Klebsiella sp. | 40–80 |
| Bifidobacterium bifidum | 30–70 |
| Staphylococcus aureus | 30–50 |
| Lactobacillus | 20–60 |
| Clostridium perfringens | 25–35 |
| Proteus mirabilis | 5–55 |
| Clostridium tetani | 1–35 |
| Clostridium septicum | 5–25 |
| Pseudomonas aeruginosa | 3–11 |
| Salmonella enteritidis | 3–7 |
| Faecalibacterium prausnitzii | ?common |
| Peptostreptococcus sp. | ?common |
| Peptococcus sp. | ?common |
The small intestine contains a trace amount of microorganisms due to the proximity and influence of the stomach. Gram positive cocci and rod shaped bacteria are the predominant microorganisms found in the small intestine.[41] However, in the distal portion of the small intestine alkaline conditions support gram-negative bacteria of the Enterobacteriaceae.[41] The bacterial flora of the small intestine aid in a wide range of intestinal functions. The bacterial flora provide regulatory signals that enable the development and utility of the gut. Overgrowth of bacteria in the small intestine can lead to intestinal failure.[43] In addition the large intestine contains the largest bacterial ecosystem in the human body.[41] Factors that disrupt the microorganism population of the large intestine include antibiotics, stress, and parasites.[41]
Bacteria make up most of the flora in the colon[44] and 60% of the dry mass of feces.[18] This fact makes feces an ideal source to test for gut flora for any tests and experiments by extracting the nucleic acid from fecal specimens, and bacterial 16S rRNA gene sequences are generated with bacterial primers. This form of testing is also often preferable to more invasive techniques, such as biopsies. Somewhere between 300[18] and 1000 different species live in the gut,[19] with most estimates at about 500.[30][32] However, it is probable that 99% of the bacteria come from about 30 or 40 species, with Faecalibacterium prausnitzii being the most common species in healthy adults.[20][45] Fungi and protozoa also make up a part of the gut flora, but little is known about their activities. The virome is mostly bacteriophages.[46]
Research suggests that the relationship between gut flora and humans is not merely commensal (a non-harmful coexistence), but rather is a mutualistic, symbiotic relationship.[19] Though people can (barely) survive with no gut flora,[30] the microorganisms perform a host of useful functions, such as fermenting unused energy substrates, training the immune system via end products of metabolism like propionate and acetate, preventing growth of harmful species, regulating the development of the gut, producing vitamins for the host (such as biotin and vitamin K), and producing hormones to direct the host to store fats.[41]: 713ff Extensive modification and imbalances of the gut microbiota and its microbiome or gene collection are associated with obesity.[47] However, in certain conditions, some species are thought to be capable of causing disease by causing infection or increasing cancer risk for the host.[18][44]
Life stages[edit]
The composition of the gut microbiota changes with age.[7][1]
Pregnancy[edit]
A study published in Cell demonstrated that a woman's gut microbiota changes as pregnancy advances. The mother's gut microbiota undergoes marked changes, with a protective effect, between the first and third trimesters of a normal, healthy pregnancy.[48]
The diversity of the flora decreases from the first to the third trimester, as the numbers of certain species rise.[49]
In addition, changes in the mother's vaginal microbiome during pregnancy are associated with effects on the infant's gut microbiota and possibly on the developing brain.[50]
A normal fetus has long been considered to be sterile; however, it is now accepted that the fetus is exposed to microbes before birth[6]. Evidence shows that there are bacteria in the amniotic fluid in utero[1] and some researchers have discovered a small community of bacteria living in the placenta.[2][51]
The maternal gut microbiome produces metabolites that can influence fetal brain development. Stress during pregnancy or maternal stress during childhood shows negative effects on the development of the brain and the gut microbiota.[52]
Early life[edit]
Mode of delivery[edit]
During birth and rapidly thereafter, bacteria from the mother and the surrounding environment colonize the infant's gut and the rest of its body.[6][22]
The first colonizers are generally aerotolerant, as the gut initially contains oxygen, and these are replaced by anaerobes typical of the adult gut microbiota[7][1], such as Escherichia coli and Streptococcus spp,[25] as well as Bifidobacteria, Clostridia and Bacteroides spp.[6]
Immediately after vaginal delivery, a baby may have bacterial strains derived from the mother's vaginal canal and feces on its body.[53]
For infants born by caesarean section (C-section), the initial exposure is most likely to be from the surrounding environment, air, the nursing staff,[54] and contact with the mother's skin.[25][1] The initial inoculum of babies born by C-section is typically dominated by the Staphyloccocus, Corynebacterium and Propionibacterium species.[55][7][1] The bacteria of infants born by emergency C-section (after initiation of labor) is closer to those born vaginally than to those born by planned C-section.[56]
Babies born by C-section have a greater risk of celiac disease, type 1 diabetes and even asthma[56][57][58], but it is not yet known whether the differences in gut microbial colonization account for this. According to some scientists, modulating early host-microbe interaction by maternal probiotic intervention during pregnancy and breastfeeding could reduce the risk of disease in children.
After birth, environmental, oral and cutaneous bacteria are readily transferred from the mother to the infant through suckling, as well as maternal kissing and caressing[5]. Early gut bacterial colonization in infants also depends on hygiene and antibiotic use[25][22][6]
Although the diversity of both bacteria and viruses in the infant gut is very low, their numbers climb over time.[7][1]
Initial colonizers are gradually replaced by phylotypes that are different from those found in the mother.[7]
Within a few days, the bacterial numbers reach 108 to 1010 per gram of infant feces.[54][59] During the first week of life, these bacteria create an environment favorable for the subsequent bacterial succession of strict anaerobic species, primarily of the genera Bifidobacterium, Bacteroides, Clostridium, and Ruminococcus.[60]
Feeding practices[edit]
Not only does breast milk contain nutrients evolutionarily programmed for human survival, it is also packed with complex carbohydrates that feed the gut microbiota.[57]
Breast-fed babies' gut microbiota becomes dominated by bifidobacteria, possibly due to the growth media for bifidobacteria in the mother's breast milk.[61][62] By contrast, the microbiota of formula-fed infants is more diverse, with high numbers of Enterobacteriaceae, enterococci, bifidobacteria, Bacteroides, and clostridia.[63][64]
Breast milk oligosaccharides nourish a particular gut bacterium called Bifidobacterium infantis, which is uniquely well suited to breaking down and making use of the specific compounds in the mother’s milk. When all goes well, the bifidobacteria proliferate and dominate, possibly helping to keep the infant healthy.[3]
The microbiome is enriched early in life in genes to facilitate lactate utilization when the infant's diet is breast milk or formula.[2] With the introduction of solid foods and the weaning off of breast milk, the types of microbes gradually shift.[3] The functional capacity to utilize plant-derived glycans is present before the introduction of solid food, suggesting that the infant gut microbes are ready to switch to a diet not exclusively based on milk before the actual change in diet takes place.[1] The weaned infant microbiota is enriched in genes for polysaccharide breakdown and vitamin production.[2]
The bacterial composition of the gut begins to converge toward an adult-like microbiota by the end of the first year of life.[1] In 2015, two papers reported that children around 10 years of age have a microbiota between adults and infants. Until then, different taxa are still developing toward an adult-type configuration.[65]
Childhood[edit]
It has been demonstrated that there are common patterns of microbiome composition evolution during life.[66] In general, the diversity of microbiota composition of fecal samples is significantly higher in adults than in children, although interpersonal differences are higher in children than in adults.[67] Much of the maturation of microbiota into an adult-like configuration happens during the three first years of life.[67]
As the microbiome composition changes, so does the composition of bacterial proteins produced in the gut. In adult microbiomes, a high prevalence of enzymes involved in fermentation, methanogenesis and the metabolism of arginine, glutamate, aspartate and lysine have been found. In contrast, in infant microbiomes the dominant enzymes are involved in cysteine metabolism and fermentation pathways.[67]
Adulthood[edit]
The diversity of gut microbiota composition is significantly higher in adults than in children.[67]
Once the gut microbiota is established, its composition remains stable until older age[Note 1].[1] However, this temporal consistency assumes that numerous variables, including diet, disease, and environment, are also held constant.[1]
Older adulthood[edit]
Both low-grade inflammation and microbial changes in the gut (decreased diversity) are characteristic of older age.[6][25]
Gut microbiota composition is also correlated with frailty. Researchers from Ireland looked at stool samples from more than 700 healthy British twins and found that a group of bacteria belonging to the species Faecalibacterium prausnitzii were found in higher amounts in the healthier twins. In the frailer twins, another group of bacteria called Eubacterium dolichum, was found.[68]
A study in 2016 showed that microbiota composition of elderly patients in long-term care varied according to their level of health dependency. Patients in long-term care settings had a less diverse microbiota than those in short-term care. Dietary intake differed significantly by residence location and appeared to drive the changes in microbiota composition.[69]
Influence of diet[edit]
Gut microbiota is influenced by the host's diet.[25] Studies show that altering diet dramatically changes the microbiota in a short time frame. Shifting to a high-fat/low-fiber "Western" diet from a low-fat, plant polysaccharide-rich diet can change the microbiota within a day.[7][25][1][29][13]
In animals,[Note 2] herbivores have more diverse gut microbiota than carnivores, indicating that degradation of plant polysaccharides is more complex and demanding on the organism.[25]
In humans, a diet rich in animal proteins, amino acids, and saturated fats is dominated by Bacteroides species. By contrast, Prevotella and Firmicutes species are associated with a diet rich in fiber,[25] carbohydrates[1][7][38] and simple sugars[38].
Malnourished human children have less mature and less diverse gut microbiota than healthy children, and changes in the microbiome associated with nutrient scarcity can in turn be a pathophysiological cause of malnutrition.[70][71] Malnourished children also typically have more potentially pathogenic gut flora, and more yeast in their mouths and throats.[72] Altering diet may lead to changes in gut microbiota composition and diversity. [73]
Dietary adjustments may allow modulation of the gut microbiota for improving health.[38] A high-fiber diet induces a microbiota composition associated with health; some argue this is partly due to increased SCFA (short-chain fatty acids) production and its anti-inflammatory effects.
Over generations of exposure to diets low in fiber, research shows, some microbial species in mice die off, decimating the diversity of the gut microbiota.[74][75][74]
Microbial diversity can be fostered by eating fermented foods such as fermented dairy and vegetables, a variety of polysaccharides, and various types of fiber, including resistant starch (bananas, oats, beans), soluble fiber (onions, root vegetables, nuts) and insoluble fiber (whole grains and avocados).[3]
Clinical and animal studies have suggested that some probiotics and prebiotics can alleviate symptoms of some gastrointestinal conditions.[76]
The gut microbiota of people in the West looks very different from that of people in other geographic locations[3], American and European guts contain relatively high levels of Bacteroides and low levels of Prevotella[24], while Prevotella dominate the guts of Africans. Researchers often attribute these differences to diet: non-Western populations may have diets with a considerable number of whole grains and plant fibers, with very little meat.[3]
Gut microbiota of different populations also appears to do specialized jobs, depending on habitual diet. For example, the gut microbiota of Japanese populations is unique in harboring an enzyme acquired from a marine bacterium that helps in the digestion of seaweed, an item prevalent in the Japanese diet.[1]
Variations in trade off of Prevotella, the representation of the urease gene, and the representation of genes encoding glutamate synthase/degradation or other enzymes involved in amino acids degradation or vitamin biosynthesis show significant differences between populations from USA, Malawi or Amerindian origin.[67]
The US population has a high representation of enzymes encoding the degradation of glutamine and enzymes involved in vitamin and lipoic acid biosynthesis; whereas Malawi and Amerindian populations have a high representation of enzymes encoding glutamate synthase and they also have an overrepresentation of α-amylase in their microbiomes. As the US population has a diet richer in fats than Amerindian or Malawian populations which have a corn-rich diet, the diet is probably a main determinant of gut bacterial composition.[67]
Further studies have indicated a large difference in the composition of microbiota between European and rural African children. The fecal bacteria of children from Florence were compared to that of children from the small rural village of Boulpon in Burkina Faso. The diet of a typical child living in this village is largely lacking in fats and animal proteins and rich in polysaccharides and plant proteins. The fecal bacteria of European children was dominated by Firmicutes and showed a marked reduction in biodiversity, while the fecal bacteria of the Boulpon children was dominated by Bacteroidetes. The increased biodiversity and different composition of gut flora in African populations may aid in the digestion of normally indigestible plant polysaccharides and also may result in a reduced incidence of non-infectious colonic diseases.[77]
Other influences[edit]
It has been shown that sharing numerous common environmental exposures in a family is a strong determinant of individual microbiome composition. This effect has no genetic influence and it is consistently observed in culturally different populations.[67]
Twin studies have shown human genes influence a subset of gut microbial species.
Exercice can also alter the gut microbiota. Several recent studies have demonstrated that exercise can modulate the gut flora in a manner that benefits health.[78]
Lately, research has also suggested that the gut microbiota of an individual may vary on different time scales, such as the time of the day.[79]
Gut microbiota functions[edit]
In experimental germ-free conditions, mice can survive without gut flora.[30] However, a germ-free existence is a life with complications. Tests have shown that a mouse raised devoid of bacteria fails to develop a proper immune system, and suffers from digestive system defects such that it must consume much more food to extract adequate calories.[23] Rodents lacking in gut flora need to eat 30% more calories just to remain the same weight as their normal counterparts.[19]
Bacteria in the gut fulfill useful functions for the host, including repressing the growth of harmful microorganisms, and training the immune system to defend the body against diseases[18][19][80].
Gut microbiota is also involved in energy harvest and storage[81], as well as metabolic functions such as breaking-down the carbohydrates that the body is not able to digest itself. Bacteria not only produce enzymes to break down certain foods,[23] but also produce vitamins that are mandatory for the body to function.[25][82] These include biotin and vitamin K.[83][84] Bacteria in the gut also aid in producing hormones to direct the host to store fats and metabolizing bile acids, sterols and xenobiotics.[85][86][18][19][30][87]
In return, the host provides these microorganisms with an oxygen-protected, nutrient-rich environment in which they can thrive.
Carbohydrate fermentation and absorption[edit]
Without gut flora, the human body would be unable to utilize some of the carbohydrates that have been consumed, because some types of gut flora have enzymes for breaking down certain polysaccharides that human cells lack.[19] Carbohydrates that humans cannot digest without bacterial help include certain starches, fibers, oligosaccharides, as well as sugars[18][32][20] such as lactose (in the case of lactose malabsorption).[32]
The majority of carbohydrate fermentation occurs in the proximal colon. When bacteria in the gut ferment carbohydrates, they produce short-chain fatty acids (SCFAs)[32][20] through a process called saccharolytic fermentation.[32] SCFAs include acetate, propionate and butyrate.[32][20] These materials can be used by host cells, providing a major source of useful energy and nutrients for humans,[32] as well as helping the body to absorb essential dietary minerals such as calcium, magnesium and iron.[18] Gases and organic acids, such as lactic acid, are also produced by saccharolytic fermentation.[20]
An estimated 10% of daily energy requirements come from colonic fermentation.[24]
The most important SCFA is butyrate, metabolised by the colonic epithelium[18] and providing energy to gut cells. Acetate is used by the muscles, while propionate helps the liver produce ATP.[32] Efficient conversion of complex indigestible dietary carbohydrates into SCFAs results in microbial cross-feeding within the community.[24] SCFAs also regulate appetite by stimulating satiety hormones,[88] and energy homeostasis[89]
Recognition of carbohydrate fermentation as a core activity of the gut microbiota provides the scientific basis for a rational design of functional foods aimed at improving gut health and also at impacting microbiota activities linked to systemic host physiology.[24]
Suppression of pathogenic microbial growth[edit]

Another important role of gut flora is preventing potentially harmful species from colonizing the gut. Gut microbiota achieves this through competitive exclusion, an activity termed the "barrier effect". Under normal circumstances, harmful yeasts and bacterial species such as Clostridium difficile are unable to grow excessively due to competition from comensal gut flora species adhering to the mucosal lining of the intestine. The barrier effect protects humans from both invading species (pathogens) and potentially harmful species normally present in the gut in low numbers (pathobionts).[18]
Commensal bacteria prevent the growth of pathogenic species by competing for nutrition and attachment sites in the epithelium of the colon. Symbiotic bacteria are more at home in this ecological niche and are thus usually more successful in competiting. Indigenous gut flora also produce bacteriocins, which are proteinaceous toxins that inhibit the growth of similar bacterial strains. These substances serve to kill pathogenic microbes[18], proving antibiotic-like actions.
Lactic acid and various fatty acids produced by bacterial fermentation also serve to lower the pH level in the colon, preventing the proliferation of harmful bacteria species. A lower pH level may also enhance the excretion of carcinogens.[32]
Immune function[edit]
This section's factual accuracy is disputed. (October 2015) |
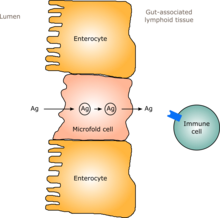
The immune system of the gut, often referred to as the gut-associated lymphoid tissue is extensive and includes the physical barrier of the intestinal wall and its mucosal coating as well as components of the innate and adaptive immune systems.
The physical barrier includes acid in the stomach, mucus and tightly connected epithelial cells, which all act to prevent the entry of pathogens.
Newborn babies have an immature immune system.[90] After birth, immunological competence is gained partly as a result of maturation factors present in breast milk and partly as a result of exposure to antigens from food and from environmental micro-organisms.
In humans, a gut flora similar to an adult's is formed within one to two years of birth.[91] As the gut flora gets established, the lining of the intestines – the intestinal epithelium and the intestinal mucosal barrier that it secretes – develop as well, in a way that is tolerant to, and even supportive of, commensurate microorganisms to a certain extent and also provides a barrier to pathogenic ones.[91] Specifically, goblet cells that produce the mucosa proliferate, and the mucosa layer thickens, providing an outside mucosal layer in which "friendly" microorganisms can anchor and feed, and an inner layer that even these organisms cannot penetrate.[91][92] Additionally, the development of gut-associated lymphoid tissue (GALT), which forms part of the intestinal epithelium and which detects and reacts to pathogens, appears and develops during the time that the gut flora develops and established.[91] The GALT that develops is tolerant to gut flora species, but not to other microorganisms.[91] GALT also normally becomes tolerant to food to which the infant is exposed, as well as digestive products of food, and gut flora's metabolites produced from food.[91]
The human immune system creates cytokines that can drive the immune system to produce inflammation in order to protect itself, and that can tamp down the immune response to maintain homeostasis and allow healing after insult or injury.[91] Different bacterial species that appear in gut flora have been shown to be able to drive the immune system to create cytokines selectively; for example Bacteroides fragilis and some Clostridia species appear to drive an anti-inflammatory response, while some segmented filamentous bacteria drive the production of inflammatory cytokines.[91][93] Gut flora can also regulate the production of antibodies by the immune system.[91][94] These cytokines and antibodies can have effects outside the gut, in the lungs and other tissues.[91]
75% of the immune system is held in the gut. It is named gut-associated lymphoid tissue (GALT).
Microbes are held in the intestinal lumen through the combined efforts of the epithelial barrier, the mucus layer, antimicrobial peptides and antibodies. They can affect important immune cells located under the epithelial surface.
The gut microbiota is essential for normal development and homeostasis of the immune system in the gut.[2]. The immune system is constantly watching and sampling the gut microbes, while the gut microbes are constantly testing the vigilance of the immune system.[57]
The intestinal microbiota stimulates the gut mucosa to produce antibodies to pathogens. Thus, the immune system learns to recognize and fight harmful bacteria and develops a tolerance for useful species.[18][30][95]
Bacteria can influence the phenomenon known as oral tolerance, in which the immune system is less sensitive to antigens (foreign substances, including those produced by gut bacteria) it encounters. The gastrointestinal immune system and the liver both partly mediate this tolerance, which can reduce an overreactive immune response such as that found in allergies and autoimmune diseases.[96] Oral tolerance is dependent on the maturation of regulatory T (Treg) cells that evolve in response to signals from the gut flora, delivered by other immune cells. Different species of the gut microbiota have different impacts on the cells of the immune system and on the probability of acquiring oral tolerance.[97] The ability of the gut flora to exert these immunomodulatory effects on the host may be restricted to a ‘critical window’ during an infant's development when the flora population is first becoming established and the immune system is completing its development.[98] The latest research supports this hypothesis by showing associations between altered patterns of microbial colonization during this critical window and an increased likelihood of developing atopic diseases.[99] Thus it appears bacterial colonization of an infant's body immediately after birth is important in modulating immune response later in life.
Gut bacteria also play a role in the manufacture of a suite of signaling molecules that communicate with and influence the immune and metabolic systems, including: neurotransmitters (including serotonin), enzymes, and vitamins.[3]
One of the questions in microbiome research is why people in modern society, who are relatively free of infectious diseases, are so prone to inflammatory, autoimmune and allergic diseases. Many scientists suspect that society-wide shifts in microbial communities have contributed to seemingly over-active immune systems. Drivers of these microbiome changes might include common use of antibiotics, sanitary practices that are aimed at limiting infectious disease but that also impact symbiotic microbes, and dietary shifts.
According to the hygienist theory, increased levels of hygiene have decreased exposures to microbes (like bacteria, parasites, fungi), and increased prevalence of both autoimmune and allergic diseases.[100][101][102] Dysbiosis can be observed in these cases or in the case of these diseases.
Metabolism and absorption[edit]
The gut microflora controls intestinal epithelial cell differentiation and proliferation through the production of SCFAs.
Gut microbiota also mediates other metabolic effects such as the syntheses of vitamins like biotin (B7) and folate (B9), as well as the absorption of ions including magnesium, calcium and iron.[31] Methanogenic archae such as Methanobrevibacter smithii are involved in the removal of end products of bacterial fermentation such as hydrogen.
Also, gut microbiota contributes to the glutathione and amino acid metabolism of the host. Glutathione is a small protein, produced inside the cells from three amino acids obtained from food or supplementation.[103] Glutathione is known to be a key antioxidant and glutathione deficiency contributes to oxidative stress, which plays a major role in several diseases and complex disorders.[103]
Neurotransmitter production[edit]
50% of the dopamine and 95% of the serotonin (the hormone of happiness) in human beings originate in the intestine,[104] where these chemical signals assist in regulating appetite and feelings of fullness and digestion.[82]
Gut-brain axis[edit]
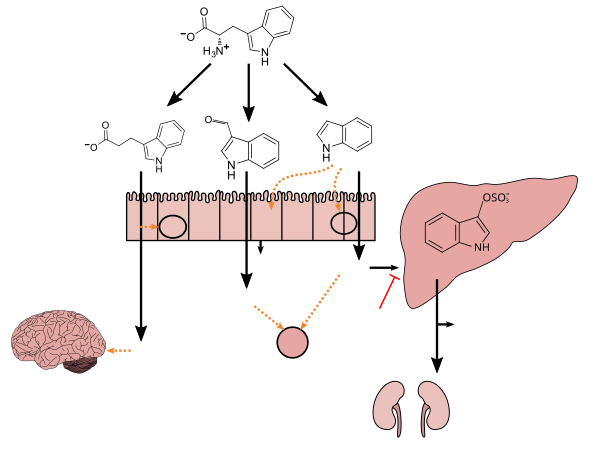
Tryptophan metabolism by human gastrointestinal microbiota ()
|
The gut–brain axis is the biochemical signaling that takes place between the gastrointestinal tract and the central nervous system.[109] That term has been expanded to include the role of the gut flora in the interplay; the term "microbiome-gut-brain axis" is sometimes used to describe paradigms explicitly including the gut flora.[109][110][111]
Broadly defined, the gut-brain axis includes the central nervous system, neuroendocrine and neuroimmune systems including the hypothalamic–pituitary–adrenal axis (HPA axis), sympathetic and parasympathetic arms of the autonomic nervous system including the enteric nervous system, the vagus nerve, and the gut microbiota.[109][111]
Interest in the field was sparked by a 2004 study showing that germ-free mice showed an exaggerated HPA axis response to stress compared to non-GF laboratory mice.[109] As of January 2016, most of the work that has been done on the role of gut flora in the gut-brain axis had been conducted in animals, or characterizing the various neuroactive compounds that gut flora can produce, and studies with humans measuring differences between people with various psychiatric and neurological differences, or changes to gut flora in response to stress, or measuring effects of various probiotics (dubbed "psychobiotics in this context), had generally been small and could not be generalized; whether changes to gut flora are a result of disease, a cause of disease, or both in any number of possible feedback loops in the gut-brain axis, remained unclear.[109][112]
A systematic review from 2016 examined the preclinical and small human trials that have been conducted with certain commercially available strains of probiotic bacteria and found that among those tested, Bifidobacterium and Lactobacillus genera (B. longum, B. breve, B. infantis, L. helveticus, L. rhamnosus, L. plantarum, and L. casei), had the most potential to be useful for certain central nervous system disorders.[113]
Disruptions in gut flora[edit]
Antibiotic use[edit]
Many antibiotics perturb the composition of the microbiota.[7] Antibiotics given to infant mice may have long-term effects on the animals’ metabolism and gut microbiota.[114]
There is now compelling evidence of major alterations of the human microbiota following treatment with antibiotics, and more frequently after repeated treatments.[1] Some taxa do not recover even months after treatment, and in general, there is a long-term decrease in bacterial diversity.[1]
By altering the population of the gastrointestinal (GI) tract microbiota, antibiotics may change the intracommunity metabolic interactions, and modify caloric intake. They may globally affect host metabolic, hormonal and immune homeostasis.[115]
Altering the numbers of gut bacteria, for example by taking broad-spectrum antibiotics, may affect the host's health and ability to digest food.[116] Changing the numbers and species of gut flora can reduce the body's ability to ferment carbohydrates and metabolize bile acids; carbohydrates that are not broken down may absorb too much water and cause runny stools.[20]
Antibiotics can cause antibiotic-associated diarrhea (AAD) by irritating the bowel directly, changing the levels of gut flora, or allowing pathogenic bacteria such as C. difficile and Salmonella kedougou to grow.[20] Another harmful effect of antibiotics is the increase of antibiotic-resistant bacteria after antibiotic use and overuse which can lead to a reduce effectiveness of ulterior treatment. Having less efficient treatments can allow bacteria to invade the host and cause illness.[116]
In a study performed on mice,[115] the ratio of Firmicutes and Lachnospiraceae was significantly elevated in animals treated with subtherapeutic doses of different antibiotics. By analyzing the caloric content of feces and the concentration of SCFAs in the GI tract, the researchers concluded that changes in the composition of microbiota lead to an increased capacity to extract calories from otherwise indigestible constituents, and to an increased production of SCFAs. These findings provide evidence that antibiotics disrupt not only the composition of the GI microbiome but also its metabolic capabilities, specifically with respect to SCFAs.[115]
Children who take certain antibiotics called macrolides to treat an infection may experience changes in their gut bacteria, and children given more than two courses of macrolides during the first two years of life appear to face an increased risk of developing asthma and becoming overweight later in childhood. Scientists do not know why, but these risks could be partly due to the early-life changes of gut microbiota composition.[117]
A number of studies have found a correlation between use of antibiotics and development of asthma or inflammatory disorders, including colorectal cancer and inflammatory bowel disease.[4] While not proven, it is hypothesized that antibiotics may deplete bacteria that favorably calibrate the immune system, leaving it prone to overreaction.[4]
When taken orally, antibiotics cause collateral damage and kill off many of the organisms within the large intestine. However, the gut microbiota show remarkable resilience and can re-attain its original composition within a short period of time once the antibiotic intervention has been removed.[118]
Other drugs[edit]
Emerging evidence shows certain drugs, such as proton pump inhibitors, affect gut microbiota composition. Xenobiotics (foreign substances and drugs) may shape the composition of the gut microbiota through antimicrobial activity or by encouraging selective bacterial growth.
Illness[edit]
Gut flora composition may change in severe illnesses, due not only to antibiotic use but also to such factors as ischemia of the gut, failure to eat, and immune compromise.[119] Negative effects from this have led to interest in selective digestive tract decontamination (SDD), a treatment to kill only pathogenic bacteria and allow the re-establishment of healthy ones.[119]
Dietary components[edit]
Under some circumstances, certain food additives (e.g. specific emulsifiers) can alter the gut microbiome and reduce the barrier layer of mucus between immune cells and bacteria, according to a study on animals published in 2015 in Nature.[120]
Some food emulsifiers also may change the profile of the gut microbiota, and thus lead to the development of low-grade inflammation in the gut, other organs and in the brain, including the appetite-control regions of the brain.[52]
Role in disease[edit]
In the last decade, the human gut microbiome has been associated with many diseases: obesity[25], inflammatory bowel syndrome, diabetes, atherosclerosis, non-alcoholic fatty liver disease, Parkinson's disease, cardiovascular disease, colon cancer, asthma, and several more.[103][1] Studies also make links between gut microbiota and brain-related disorders such as Alzheimer's disease, autism, depression, anxiety, and schizophrenia.[23][2] To date, none of these conditions are known to be caused exclusively by gut microbiota. More details on these connections will become clearer as research progress.
Ulcers[edit]
Helicobacter pylori can cause stomach ulcers by crossing the epithelial lining of the stomach. Here the body produces an immune response. During this response parietal cells are stimulated and release extra hydrochloric acid (HCl+) into the stomach. However, the response does not stimulate the mucus-secreting cells that protect and line the epithelium of the stomach. The extra acid sears holes into the epithelial lining of the stomach, resulting in stomach ulcers.[66]
Cancer[edit]
Colorectal cancer (CRC) is known to be connected with a Western lifestyle and in particular with a diet high in meat and fat and low in fiber. Accordingly, colon cancer rates are much higher in Western countries than in countries located in Africa or the Far East.[29]
While genetic risk is a factor in CRC, there is evidence from global studies that the microbiome plays a role in the level of cancer risk.[24]
Some genera of bacteria, such as Bacteroides and Clostridium, have been associated with an increase in tumor growth rates, while other genera, such as Lactobacillus and Bifidobacteria, are known to prevent tumor formation.[18] Metagenomic analyses have also identified a reduction of butyrate-producers in patients with colorectal cancer.[24]
Gut bacteria thus appear to be a promising target in developing measures to prevent and treat colon cancer.[29]
Gut microbes can also influence the efficacy of cancer therapy.[121] According to two studies from independent research teams published in 2015 in Science magazine, the presence of certain types of gut microbes in mice can boost the anti-tumor effects of cancer immunotherapy.[121]
Inflammatory bowel disease[edit]
The incidence of inflammatory bowel disease (IBD) has risen rapidly over the last several years. Crohn’s disease and ulcerative colitis are the main IBDs and are characterized by a chronic and exacerbated inflammation of the intestinal mucosa.[2]
IBD is more common in people who were not breastfed as infants, those who had good hygiene in youth, and those with a poor diet.[122] Its incidence is inversely linked with poor sanitation during the first years of life and consumption of fruits, vegetables, and unprocessed foods.[122] Also, the use of antibiotics, especially during childhood, is associated with IBD.[81] Some believe these factors support the idea gut microbiota is involved in the disease.
It has been noted that though ulcerative colitis and Crohn's disease have genetic components, they are probably due to a complex set of factors rather than solely to genetics.[123] Both lifestyle factors and an altered gut microbiota may contribute to IBDs.[123][2]
Changes in the gut microbiota have been repeatedly reported in patients with IBD: in general, patients with IBD, compared to healthy controls,[24] tend to have fewer bacteria with anti-inflammatory properties and/or more bacteria with proinflammatory properties. In addition, alterations in the two dominant phyla (Firmicutes and Bacteroidetes) are observed, coupled with an increase in abundance of members of the Proteobacteria phylum.[24] There may be spatial reorganization of the Bacteroides species in patients with IBD, with Bacteroides fragilis being responsible for a greater proportion of the bacterial mass in patients with IBD compared with controls.
Specifically, a reduction in Faecalibacterium prausnitzii and increased numbers of Escherichia coli have been documented in patients with Crohn's disease.[24] Furthermore, some bacterial strains such as Clostridium difficile[122] are associated with colitis.[124]
It has also been noted that the IBD metagenome contains 25% fewer genes than the healthy gut, with studies showing a correlative decrease in proteins and functional pathways.[24]
Emerging research shows commensal microbiota may play an important role in the pathogenesis of IBD.[2] Scientists recently identified a type of regulatory T cell that is unique to humans and deficient in people with inflammatory bowel disease.[4] Two recent papers published in Nature showed that butyrate is important in enlisting regulatory T cells, a branch of immune cells that control the processes involved in IBD.[23]
Butyrate, which is produced by bacteria of the Bacteroides and Firmicutes phyla after fermentation of dietary fiber, shows anti-inflammatory properties in individuals with IBD. Patients with colitis and Crohn's disease have reduced levels of these bacteria in the colon. A study published in 2016 shows that fungi could be at the origin and cause Crohn's disease.[125]
Abnormal tight junctions, which are supposed to prevent gut permeability, have been found in those with IBD.[126] However, it is not clear whether inflammation causes the increased intestinal permeability or vice versa.[126]
Several clinical trials have examined the approach of modulating the microbiota on patients with IBD through fecal microbiota transplantation.[24] In ulcerative colitis, the strategy is moderately promising.[24]
Some studies also suggest certain probiotic strains may also help symptoms of mild to moderate ulcerative colitis.[127]
The two main types of inflammatory bowel diseases, Crohn's disease and ulcerative colitis, are chronic inflammatory disorders of the gut; the causes of these disease are unknown and issues with the gut flora and its relationship with the host have been implicated in these conditions.[128][129][130][131] Additionally, it appears that interactions of gut flora with the gut-brain axis have a role in IBD, with physiological stress mediated through the hypothalamic–pituitary–adrenal axis driving changes to intestinal epithelium and the gut flora in turn releasing factors and metabolites that trigger signaling in the enteric nervous system and the vagus nerve.[132]
The diversity of gut flora appears to be significantly diminished in people with inflammatory bowel diseases compared to healthy people; additionally, in people with ulcerative colitis, Proteobacteria and Actinobacteria appear to dominate; in people with Crohn's, Enterococcus faecium and several Proteobacteria appear to be over-represented.[132]
There is reasonable evidence that correcting gut flora imbalances by taking probiotics with Lactobacilli and Bifidobacteria can reduce visceral pain and gut inflammation in IBD.[112]
Obesity[edit]
According to the "missing microbiota hypothesis" of Martin J. Blaser, impoverishment of the gut microbiota over successive generations may be contributing to the epidemic of obesity.[3][133]
Obesity is associated with changes in the composition of the gut microbiota. An increased ratio of Firmicutes to Bacteroidetes has been observed in the microbiota of some obese individuals,[25] with weight loss associated with increasing levels of Bacteroidetes,[24][134] although this pattern is not always observed.
Some scientists believe the influence of gut flora composition on weight is attributable to differences in the energy absorbing potential of different ratios of Firmicutes to Bacteroidetes. The digestion of fatty acids and dietary polysaccharides may differ with this ratio, as shown by experiments[135] in which the gut flora of obese mice were transplanted into germ-free recipient mice, leading to an increase in weight despite a decrease in food consumption.[136][137][138][139][140] Studies in which microbes from either an obese mouse or an obese human are implanted into a germ-free mouse show an increase in the lean animals’ body weight,[141] suggesting a causal relationship between the altered microbiota and obesity development.[25] While most scientists agree that gut microbiota play a role in human obesity, some studies are contradictory.[21]
Chronic low-grade inflammation, modulation of secretion of gut-derived peptide hormones, regulation of active adipose tissue composition, and increased energy harvest from host diet have all been suggested as mechanisms through which the gut microbiota may contribute to obesity.[21]
According to a recent research, done on a Mexican population, which purpose was to investigate the relationship between the gut microbiota composition, the lipopolysaccharide (a component of the bacterial cell wall) levels in the blood and the metabolic profile in obese and normal-weight young subjects, high metabolic endotoxemia (determined by serum concentration of lipopolysaccharides) was related to a smallest amount of Escherichia coli, high triglyceride levels, and central adiposity in obese young persons.
In other words, a disruption in the types of gut microorganisms (dysbiosis) can lead to an effect called “metabolic endotoxemia”, which is connected to weight gain and insulin resistance.[142]
The low-grade inflammation and insulin resistance observed in obesity may be triggered by alteration of the gut barrier, leading to the higher plasma lipopolysaccharide (LPS) levels observed in obese individuals.[2]
It is also possible that lower diversity in the gut microbiota may have an effect on satiety and eating behavior.[21] The gut microbiome affects energy expenditure and storage in the host[2] by producing signaling chemicals that regulate appetite, satiety and digestion.[3]
The early life microbiota may be important in later obesity. Children who are overweight may have higher levels of Staphylococcus aureus and lower levels of Bifidobacteria during infancy.[25] In addition, mice that have their gut microbiota depleted by antibiotic treatment show a long-term impact on acquired obesity and weight gain.[21]
Dietary factors may modulate the gut microbiota in a way that influences obesity. Recent studies have identified a high fat diet, as a strong modulator of the microbiota.[24] An increased intake of dietary fiber, which is fermented in the colon, has been reported to decrease body weight and glucose control.[2]
A study in the European Journal of Clinical Nutrition found that people who drank fermented milk with a particular strain of Lactobacillus gasseri for 12 weeks had a reduction in abdominal fat and body weight, compared to those who consumed a control drink. Another study published in the Journal of Functional Foods, found that people who consumed yogurt containing two new strains of probiotics experienced small losses in body fat.[127]
To date, bariatric surgery, indicated for severe obesity, is the only treatment that enables substantial and sustained weight loss. It is not known how bariatric surgery improves results in weight loss, but some suggest its effects could be partly attributable to the drastic alterations it causes in gut microbiota composition.[143]
Diabetes[edit]
Many studies suggest that development of both type 1 and 2 diabetes are linked to changes in gut microbiota.[25]
The gut microbiota in children with type 1 diabetes differs from that in healthy children.[144][25] In adults with type 1 diabetes, the significant differences in the number of Bifidobacterium, Lactobacillus, and Clostridium, and in the Firmicutes to Bacteroidetes ratio observed may be related to glycemic level.[144] Furthermore, in mice predisposed to type 1 diabetes, it has been shown that the interaction of the gut microbiota with the innate immune system modifies predisposition toward developing diabetes.[1]
Children at high genetic risk for type 2 diabetes (T2D) exhibit a distinct composition of the gut microbiota, with decreased diversity over time and higher relative abundance of Bacteroides ovatus and Firmicutes strain CO19.[1]
According to other studies, butyrate-producing bacteria Roseburia spp. and Faecalibacterium spp., known to be anti-inflammatory, are less abundant in adults with T2D than in healthy control subjects.[25]
A more direct link between an altered gut microbiota and insulin resistance in humans was recently discovered. Insulin sensitivity and levels of butyrate-producing bacteria increased in patients with insulin resistance after transplantation of intestinal microbiota from lean, healthy donors.[25]
Moreover, microbial changes that occur prior to the onset of T2D may be used for early diagnosis.[145]
[edit]
Many studies demonstrate the link between gut microbiota composition and behavior and cognitive functioning.[104] The intestines are responsive to shifts in emotions and mental states,[146][23] while the human brain constantly monitors gut activity. Gut microbiota communicates with the nervous system using some of the same neurochemicals that relay messages in the brain.[82] The microbiome produces the mood-influencing neurotransmitters like serotonin, norepinephrine, and dopamine. Bacteria can also change how the central nervous system uses these chemicals.[147]
This communication could have an influence on human behavior.[82] The vagus nerve plays a major role in enabling signals to travel from brain to gut and vice versa.[148] The involved system of communication has been called the microbiota-gut-brain axis.[104]
Emerging research suggests gut microbiota is implicated in brain-related conditions that include anxiety and depression, Alzheimer’s disease and dementia[23], Parkinson’s disease, and autism.[23][104] Anxiety and depression have been linked to lower levels of Lactobacillus helveticus and Bifidobacterium longum bacterial strains, for example.[149] Also, several recent studies show the gut microbiota is altered in those with Parkinson's disease.[23]
Transplantation of the fecal microbiome from one mouse strain displaying a certain set of behaviors to another strain resulted in the recipient strain exhibiting the behavioral phenotype of the donor.[104]
Other studies in mice show that the introduction of a single, unique bacterium in the gut can result in the development of anxiety-like behavior with concomitant activation of neuronal regions in the brain, dependent on information received from the gut via the vagus nerve.[104] Researchers in one recent study took stool samples from people with depression and transplanted them into the digestive tracts of rats. The formerly carefree rodents soon began showing signs of depression and anxiety.[147]
The intestinal microbiota may even shape stroke outcome: in mice, researchers found that bacteria influenced neural activity after stroke by altering the action of gut immune cells.[150]
In 2011, a paper published in Bioessays, proposed that probiotic bacteria could be tailored to treat specific psychological diseases.[82] These potentially mind-altering microbes have been called "psychobiotics".[82]
According to a UCLA study published in 2013, modulating gut bacteria through diet can affect brain function, even in healthy individuals. Brain response was gauged by functional MRI. Findings indicated that a probiotic yogurt was able to change the way the brain responded to emotionally-charged stimuli.[151] The study highlighted the benefits of some probiotics on emotional reactivity.[151] A different study found, after taking the Bifidobacterium longum probiotic for a month, healthy people reported feeling less stress than when they took a placebo. They also had lower levels of the stress-related hormone cortisol. In addition, the subjects showed improvements on a test of visual memory.[147] These studies support the notion that diet-based therapies may be designed to influence brain function and behavior.[104]
According to a recent study, Parkinson’s disease begins in the gut and moves up the vagus nerve and into the brain.[152] Bacteria are capable of producing substances that either directly or indirectly impact brain function, through the vagus nerve, which directly connects to the brain.[152]
Asthma and allergies[edit]
Asthma prevalence is increasing in Western countries.[2] Children with asthma have a different intestinal microbiota compared to nonasthmatic children. Asthmatic children have a high prevalence of certain species of Clostridium difficile (bacterium with pathogenic characteristics) and low Bifidobacterium (nonpathogenic bacteria) in their intestinal microbiota.[2]
Bacteria may also be implicated in the development of allergies,[86] which are overreactions of the immune system to harmless antigens. Studies on the gut flora of infants and young children have shown that those who have or later develop allergies show different compositions of gut flora and higher chances of having the harmful species Clostridium difficile and Staphylococcus aureus and lower chances of having Bacteroides and Bifidobacteria than children without allergies.[86]
One explanation for the link between gut microbiota and allergies is that gut microbiota stimulate the immune system and "train" it to respond properly to antigens, so a lack of these bacteria in early life leads to an inadequately trained immune system that overreacts to specific antigens.[86] On the other hand, the differences in flora could be a consequence, not a cause, of the allergies.[86]
Modulating the immune system with probiotics is a promising strategy for prevention and treatment of allergies. In a study published in 2016, researchers showed that the introduction of the probiotic Bifidobacterium longum (Lactobacillus) was able to reduce the effects of food allergies.[153] Lactobacillus casei in yogurt may decrease body levels of immune substances involved in seasonal allergies.[154]
Meanwhile, clinical trials have indicated that feeding Lactobacillus rhamnosus GG and Lactobacillus fermentum to mothers in the prenatal and early postnatal periods may be effective in the prevention of early atopic disease in children.[2]
Moreover, the hygiene hypothesis suggests that an overly clean living environment decreases gut microbes diversity, and thus increases the incidence of asthma, inflammation, and autism.[155]
Liver disease[edit]
It is now recognized that the gut microbiota and chronic liver diseases are closely linked.
As the liver is fed directly by the portal vein, whatever crosses the intestinal epithelium and the intestinal mucosal barrier enters the liver, as do cytokines generated there.[156] Dysbiosis in the gut flora has been linked with the development of cirrhosis and non-alcoholic fatty liver disease.[156]
In the 1980s, the development of non-alcoholic steatohepatitis (NASH) and small intestinal bacterial overgrowth was observed in humans after intestinal bypass.[24]
Subsequently, a lower proportion of Ruminococcaceae has been found in patients with NASH compared with healthy subjects.[24] A study which characterized the gut microbiota of children with NASH and obesity showed that those with NASH had a higher proportion of Escherichia compared with other groups.[24]
Regression of hepatic steatosis can be observed after metronidazole treatment, suggesting a possible role for the gut bacteria in non-alcoholic fatty liver disease (NAFLD).[24] Patients with NAFLD also have increased intestinal permeability.[24]
The gut can also become more permeable in diseases like cirrhosis.[157][158] Increased gut permeability can allow translocation of bacteria through the gut mucosal lining, which is the border between the lumen of the gut and the inside of the body.[30][126] Bacteria can be very harmful to the host if they get outside of the intestinal tract.[19][26] Translocation of bacteria or microbe derived products into the portal circulation contributes to the pathogenesis of liver diseases.[24]
According to a recent study, gut microbiota alterations can predict hospitalization of patients with cirrhosis.
Probiotics have shown promise in ameliorating liver injury by reducing bacterial translocation and hepatic inflammation.[24]
Other diseases[edit]
Gut microbiota composition is linked with other diseases, like multiple sclerosis, and rheumatoid arthritis,[1] although a causal role for bacteria on these conditions has not been established.
Gut flora could also impact severe immunodeficiency in HIV disease. Two thirds of all T cells reside in the lymphoid tissue of the gut, where the human immunodeficiency virus spreads after exposure, even before it shows up in blood.[4] Further research will illuminate whether microbiota interventions could stop HIV infection from progressing to AIDS.
Irritable bowel syndrome is a result of stress and chronic activation of the HPA axis; its symptoms include abdominal pain, changes in bowel movements, and an increase in proinflammatory cytokines. Overall, studies have found that the luminal and mucosal microbiota are changed in irritable bowel syndrome individuals, and these changes can relate to the type of irritation such as diarrhea or constipation. Also, there is a decrease in the diversity of the microbiome with low levels of fecal Lactobacilli and Bifidobacteria, high levels of facultative anaerobic bacteria such as Escherichia coli, and increased ratios of Firmicutes : Bacteroidetes.[111]
In irritable bowel syndrome, the gut microbiota appears to play a role in modulating responses to visceral pain. These interactions occur at the level of the gastrointestinal mucosa.[159] Moreover, one study reports a decrease in visceral sensitivity by changing gut microbiota with probiotics in rats.
Therapeutics[edit]
Scientists aim to develop novel therapeutics and strategies to modulate the microbiota to treat or prevent disease. Additionally, in some instances it may be possible to survey the microbiome in order to detect diseases before conventional diagnostics can, and thus provide more proactive treatment.[24]
Pharmabiotics is a generic term to encompass any form of therapeutic that exploits the commensal flora, including live probiotic bacteria, probiotic-derived biologically active metabolites, prebiotics, synbiotics and genetically modified commensal bacteria.[31]
Probiotics[edit]
According to the World Health Organization, probiotics are "live microorganisms which when administered in adequate amounts confer a health benefit on the host".[160]
The use of probiotics is promising for improving health in individuals with and without disease.
There is some evidence that treatment with some probiotic strains of bacteria may be effective in irritable bowel syndrome and chronic idiopathic constipation. Those organisms most likely to result in a decrease of symptoms have included:
- Streptococcus faecium
- Lactobacillus plantarum
- Lactobacillus rhamnosus
- Propionibacterium freudenreichii
- Bifidobacterium breve
- Lactobacillus reuteri
- Lactobacillus salivarius
- Bifidobacterium infantis
- Streptococcus thermophilus[161][162][163]
A high level of evidence shows that probiotics reduce the risk of antibiotic-associated diarrhea. A Health Canada monograph stated that products containing certain probiotics, such as Lactobacillus rhamnosus GG, help manage acute infectious diarrhea and antibiotic-associated diarrhea.[127] Another review of 23 trials also concluded that probiotics are effective in preventing antibiotic-related diarrhea.[127]
Another paper published in 2011 revealed that probiotics play a major role in the prevention of antibiotic-associated diarrhea and Clostridium difficile infection.[164]
A paper discussing five controlled studies, published in the World Journal of Gastroenterology, stated that certain strains of Bifidobacterium lactis and Lactobacillus casei improved stool consistency and frequency of bowel movements in people with constipation.[127]
There is reasonable evidence that taking probiotics containing Lactobacillus species may help prevent antibiotic-associated diarrhea and that taking probiotics with Saccharomyces (e.g., Saccharomyces boulardii ) may help to prevent Clostridium difficile infection following systemic antibiotic treatment.[112]
There is also evidence supporting a therapeutic role for probiotics in treating the symptoms of irritable bowel syndrome[165] and for non-alcoholic fatty liver disease (NASH).
A reference guide published in 2013 by the European Society for Primary Care Gastroenterology (ESPCG) provides the first practical consensus on the role of specific probiotics in the management of lower GI symptoms in adults in clinical practice.[166]
Prebiotics[edit]
Prebiotics are dietary components that help foster the growth of beneficial microorganisms in the gut, leading to better health.[122]
The effect of prebiotics is to stimulate activity of "good bacteria" such as Bifidobacteria and Lactobacilli in the gut, and thereby increase the body's natural resistance to invading pathogens.[167]
Synbiotics[edit]
Synbiotics are combinations of probiotics and prebiotics, delivering both the live microorganisms and the substrate(s) promoting their growth.
On animals, synbiotics appear to be a promising antibiotic alternative.[168]
Fecal microbiota transplantation[edit]
Fecal microbiota transplant (FMT) has recently emerged as a powerful way to treat one specific disease.[24][147][57][1][4] FMT, which involves transferring a complete gut microbial community from a healthy person to a sick person', has been shown to effectively treat recurrent cases of infection by antibiotic-resistant intestinal pathogen Clostridium difficile, which kills 14,000 Americans each year.[3]
Scientists have shown how, in the majority of cases, transplanting the microbial community from a healthy donor into a patient suffering from recurrent Clostridium difficile-associated disease (CDAD) significantly modified the bacterial composition of the patient.[1] More than 81% of patients were cured of symptoms after the treatment, with few side effects.[57][4]
After 2 weeks, the microbiota of the recipient had dramatically changed to a community similar to that of the donor.
FMT is being explored as a way to address other microbiota-linked conditions, with limited success to date.
Other animals[edit]
Aside from mammals, some insects also possess complex and diverse gut microbiota that play key nutritional roles.[169] Microbial communities associated termites can constitute a majority of the weight of the individuals and perform important roles in the digestion of lignocellulose and nitrogen fixation.[170] These communities are host-specific, and closely related insect species share comparable similarities in gut microbiota composition.[171][172] In cockroaches, gut microbiota have been shown to assemble in a deterministic fashion, irrespective of the inoculum;[173] the reason for this host-specific assembly remains unclear. Bacterial communities associated with insects like termites and cockroaches are determined by a combination of forces, primarily diet, but there is some indication that host phylogeny may also be playing a role in the selection of lineages.[171][172]
For more than 51 years we have known that the administration of low doses of antibacterial agents promotes the growth of farm animals to increase weight gain.[115]
In a study performed on mice by Ilseung Cho,[115] the ratio of Firmicutes and Lachnospiraceae was significantly elevated in animals treated with subtherapeutic doses of different antibiotics. By analyzing the caloric content of faeces and the concentration of small chain fatty acids (SCFAs) in the GI tract, they concluded that the changes in the composition of microbiota lead to an increased capacity to extract calories from otherwise indigestible constituents, and to an increased production of SCFAs. These findings provide evidence that antibiotics perturb not only the composition of the GI microbiome but also its metabolic capabilities, specifically with respect to SCFAs.[115]
See also[edit]
- Human microbiome
- Human microbiome project
- List of human flora
- Microbiome
- Probiotic
- Skin flora
- Fecal microbiota transplant
Notes[edit]
Sources[edit]
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z "The Impact of the Gut Microbiota on Human Health". Cell. 2012.
- ^ a b c d e f g h i j k l m n o p q r "The Central Role of the Gut Microbiota in Chronic Inflammatory Diseases". Journal of Immunology Research. 2014.
- ^ a b c d e f g h i j k l m "Some of My Best Friends Are Germs". New York Times. 2013.
- ^ a b c d e f g "Among Trillions of Microbes in the Gut, a Few Are Special". Scientificamerican.com. 2015.
- ^ a b c d e f g "Gut microbiota: Married to our gut microbiota". Nature. 2012.
- ^ a b c d e f g h i j k l Guinane, C. M; Cotter, P. D (2013). "Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ". Therapeutic Advances in Gastroenterology. 6 (4): 295–308. doi:10.1177/1756283X13482996. PMC 3667473. PMID 23814609. Cite error: The named reference "ncbi.nlm.nih.gov" was defined multiple times with different content (see the help page).
- ^ a b c d e f g h i j k l m n o "The Impact of the Gut Microbiota on Human Health". Cell. 2012.
- ^ Qin, Junjie; Li, Ruiqiang; Raes, Jeroen; Arumugam, Manimozhiyan; Burgdorf, Kristoffer Solvsten; Manichanh, Chaysavanh; Nielsen, Trine; Pons, Nicolas; Levenez, Florence; Yamada, Takuji; Mende, Daniel R.; Li, Junhua; Xu, Junming; Li, Shaochuan; Li, Dongfang; Cao, Jianjun; Wang, Bo; Liang, Huiqing; Zheng, Huisong; Xie, Yinlong; Tap, Julien; Lepage, Patricia; Bertalan, Marcelo; Batto, Jean-Michel; Hansen, Torben; Le Paslier, Denis; Linneberg, Allan; Nielsen, H. Bjørn; Pelletier, Eric; Renault, Pierre (2010). "A human gut microbial gene catalogue established by metagenomic sequencing". Nature. 464 (7285): 59–65. doi:10.1038/nature08821. PMC 3779803. PMID 20203603.
- ^ a b "Nature review". Nature.
- ^ "The microbiome: an opportunity for smarter cosmetics". Premiumbeautynews.com. 2015.
- ^ a b "Marked seasonal variation in the wild mouse gut microbiota". The ISME Journal. 2015.
- ^ Stephen, A. M.; Cummings, J. H. (1980). "The Microbial Contribution to Human Faecal Mass". Journal of Medical Microbiology. 13 (1): 45–56. doi:10.1099/00222615-13-1-45. PMID 7359576.
- ^ a b "You Are What You Eat, All 100 Trillion Of You". National Geographic. 2013.
- ^ "5 Ways Gut Bacteria Affect Your Health". Livescience.com. 2013.
- ^ "Germs, humans and numbers: New estimate revises our microbiome numbers downwards". Science daily. 2016.
- ^ "A framework for human microbiome research". Nature. 2012.
- ^ Tap, Julien; Mondot, Stanislas; Levenez, Florence; Pelletier, Eric; Caron, Christophe; Furet, Jean-Pierre; Ugarte, Edgardo; Muñoz-Tamayo, Rafael; Paslier, Denis L. E.; Nalin, Renaud; Dore, Joel; Leclerc, Marion (2009). "Towards the human intestinal microbiota phylogenetic core". Environmental Microbiology. 11 (10): 2574–84. doi:10.1111/j.1462-2920.2009.01982.x. PMID 19601958.
- ^ a b c d e f g h i j k l m n o p q r Guarner, F; Malagelada, J (2003). "Gut flora in health and disease". The Lancet. 361 (9356): 512–9. doi:10.1016/S0140-6736(03)12489-0. PMID 12583961.
- ^ a b c d e f g h i j k Sears, Cynthia L. (2005). "A dynamic partnership: Celebrating our gut flora". Anaerobe. 11 (5): 247–51. doi:10.1016/j.anaerobe.2005.05.001. PMID 16701579.
- ^ a b c d e f g h i j k Cite error: The named reference
Beaugerie L and Petit JCwas invoked but never defined (see the help page). - ^ a b c d e f g "Does the Gut Microbiota Contribute to Obesity? Going beyond the Gut Feeling". MDPI. 2015.
- ^ a b c Geuking, Markus B; Köller, Yasmin; Rupp, Sandra; McCoy, Kathy D (2014). "The interplay between the gut microbiota and the immune system". Taylor and Francis Online. 5 (3): 411–418. doi:10.4161/gmic.29330.
- ^ a b c d e f g h i j k l m n o "I had the bacteria in my gut analysed. And this may be the future of medicine". The Guardian. 2014.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y "The gut microbiota and host health: a new clinical frontier". Gut journal. 2015.
- ^ a b c d e f g h i j k l m n o p q r s t "Assessing the Human Gut Microbiota in Metabolic Diseases". American diabetes association. 2013.
- ^ a b c Vedantam, Gayatri; Hecht, David W (2003). "Antibiotics and anaerobes of gut origin". Current Opinion in Microbiology. 6 (5): 457–61. doi:10.1016/j.mib.2003.09.006. PMID 14572537.
- ^ Segal, Leopoldo; Blaser, Martin (2014). "A Brave New World: The Lung Microbiota in an Era of Change". Annals of the American Thoracic Society. 11: S21–S27. doi:10.1513/AnnalsATS.201306-189MG. Retrieved 2014-09-14.
- ^ "The gut microbiota in IBD". Nature. 2012.
- ^ a b c d "Reducing colon cancer risk: how diet switch changes gut microbial metabolism". Medical News Today. 2016.
- ^ a b c d e f g Steinhoff, U (2005). "Who controls the crowd? New findings and old questions about the intestinal microflora". Immunology Letters. 99 (1): 12–6. doi:10.1016/j.imlet.2004.12.013. PMID 15894105.
- ^ a b c d O'Hara, Ann M; Shanahan, Fergus (2006). "The gut flora as a forgotten organ". EMBO Reports. 7 (7): 688–93. doi:10.1038/sj.embor.7400731. PMC 1500832. PMID 16819463.
- ^ a b c d e f g h i j Gibson, Glenn R. (2004). "Fibre and effects on probiotics (the prebiotic concept)". Clinical Nutrition Supplements. 1 (2): 25–31. doi:10.1016/j.clnu.2004.09.005.
- ^ Khanna S, Tosh PK (January 2014). "A clinician's primer on the role of the microbiome in human health and disease". Mayo Clin. Proc. 89 (1): 107–14. doi:10.1016/j.mayocp.2013.10.011. PMID 24388028.
- ^ "Top 20 core bacterial genera in the mouse and human gut microbiota". Nature. 2015.
- ^ a b Cui L, Morris A, Ghedin E (July 2013). "The human mycobiome in health and disease". Genome Med. 5 (7): 63. doi:10.1186/gm467. PMC 3978422. PMID 23899327.
Figure 2: Distribution of fungal genera in different body sites
{{cite journal}}: External link in|quote= - ^ Erdogan A, Rao SS (April 2015). "Small intestinal fungal overgrowth". Curr Gastroenterol Rep. 17 (4): 16. doi:10.1007/s11894-015-0436-2. PMID 25786900.
- ^ a b Arumugam, Manimozhiyan; Raes, Jeroen; Pelletier, Eric; Le Paslier, Denis; Yamada, Takuji; Mende, Daniel R.; Fernandes, Gabriel R.; Tap, Julien; Bruls, Thomas; Batto, Jean-Michel; Bertalan, Marcelo; Borruel, Natalia; Casellas, Francesc; Fernandez, Leyden; Gautier, Laurent; Hansen, Torben; Hattori, Masahira; Hayashi, Tetsuya; Kleerebezem, Michiel; Kurokawa, Ken; Leclerc, Marion; Levenez, Florence; Manichanh, Chaysavanh; Nielsen, H. Bjørn; Nielsen, Trine; Pons, Nicolas; Poulain, Julie; Qin, Junjie; Sicheritz-Ponten, Thomas; Tims, Sebastian (2011). "Enterotypes of the human gut microbiome". Nature. 473 (7346): 174–80. doi:10.1038/nature09944. PMC 3728647. PMID 21508958.
- ^ a b c d Wu, G. D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S. A.; Bewtra, M.; Knights, D.; Walters, W. A.; Knight, R.; Sinha, R.; Gilroy, E.; Gupta, K.; Baldassano, R.; Nessel, L.; Li, H.; Bushman, F. D.; Lewis, J. D. (2011). "Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes". Science. 334 (6052): 105–8. doi:10.1126/science.1208344. PMC 3368382. PMID 21885731.
- ^ "Gut microbiota in 2015: Prevotella in the gut: choose carefully". Nature Reviews Gastroenterology & Hepatology. 2016.
- ^ Zimmer, Carl (April 20, 2011). "Bacteria Divide People Into 3 Types, Scientists Say". The New York Times. Retrieved April 21, 2011.
a group of scientists now report just three distinct ecosystems in the guts of people they have studied.
- ^ a b c d e f g Cite error: The named reference
Prescottswas invoked but never defined (see the help page). - ^ Kenneth Todar (2012). "The Normal Bacterial Flora of Humans". Todar's Online Textbook of Bacteriology. Retrieved June 25, 2016.
- ^ Quigley, Eamonn M M; Rodrigo Quera (February 2006). "Small intestinal bacterial overgrowth: roles of antibiotics, prebiotics, and probiotics". Gastroenterology. 130 (2 Suppl 1): S78–90. doi:10.1053/j.gastro.2005.11.046. ISSN 0016-5085. PMID 16473077.
- ^ a b University of Glasgow. 2005. The normal gut flora. Available through web archive. Accessed May 22, 2008
- ^ Miquel, S; Martín, R; Rossi, O; Bermúdez-Humarán, LG; Chatel, JM; Sokol, H; Thomas, M; Wells, JM; Langella, P (2013). "Faecalibacterium prausnitzii and human intestinal health". Current Opinion in Microbiology. 16 (3): 255–61. doi:10.1016/j.mib.2013.06.003. PMID 23831042.
- ^ Scarpellini E, Ianiro G, Attili F, Bassanelli C, De Santis A, Gasbarrini A (2015). "The human gut microbiota and virome: Potential therapeutic implications". Dig Liver Dis (Review). 47 (12): 1007–12. doi:10.1016/j.dld.2015.07.008. PMID 26257129.
- ^ Ley Ruth E (2010). "Obesity and the Human Microbiome". Current Opinion in Gastroenterology. 26 (1): 5–11. doi:10.1097/MOG.0b013e328333d751. PMID 19901833.
- ^ "Gut microbiota: The gut microbiota is profoundly altered over the course of pregnancy". Nature. 2012.
- ^ Baker, Monya (2012). "Pregnancy alters resident gut microbes". Nature. doi:10.1038/nature.2012.11118.
- ^ "Maternal stress alters offspring gut, brain through vaginal microbiome". Endocrine Society. 2015.
- ^ Collado, M and Bӓuerl C et al. Defining microbiota for developing new probiotics. Microb Ecol Health Dis.2012;23 PMCID:PMC 3747743
- ^ a b Mayer, Emeran (2016). "The mind-gut connection". 310.
{{cite journal}}: Cite journal requires|journal=(help) - ^ Bettelheim, K. A.; Breadon, Alwena; Faiers, Mary C.; O'Farrell, Sheila M.; Shooter, R. A. (2009). "The origin of O serotypes of Escherichia coli in babies after normal delivery". Journal of Hygiene. 72 (1): 67–70. doi:10.1017/S0022172400023226. PMC 2130250. PMID 4593741.
- ^ a b Schwiertz, Andreas; Gruhl, Bärbel; Löbnitz, Manuela; Michel, Peter; Radke, Michael; Blaut, Michael (2003). "Development of the Intestinal Bacterial Composition in Hospitalized Preterm Infants in Comparison with Breast-Fed, Full-Term Infants". Pediatric Research. 54 (3): 393–9. doi:10.1203/01.PDR.0000078274.74607.7A. PMID 12788986.
- ^ Dominguez-Bello, Maria; Blaser, Martin; Ley, Ruth; Knight, Rob (2010). "Development of the Human Gastrointestinal Microbiota and Insights From High-Throughput Sequencing". Gastroenterology. 140 (6): 1713–1719. doi:10.1053/j.gastro.2011.02.011.
- ^ a b "How to keep your gut bacteria in good form". Sundayworld. 2016.
- ^ a b c d e "Gut health: Stanford researchers explain how what's in our bellies affects myriad health issues". San Jose Mercury News. 2016.
- ^ "Mom's microbes influence her offspring's immune system, mice study shows". American Association for the Advancement of Science. 2016.
- ^ MacKie, RI; Sghir, A; Gaskins, HR (1999). "Developmental microbial ecology of the neonatal gastrointestinal tract". The American Journal of Clinical Nutrition. 69 (5): 1035S–1045S. PMID 10232646.
- ^ Favier, C. F.; Vaughan, E. E.; De Vos, W. M.; Akkermans, A. D. L. (2002). "Molecular Monitoring of Succession of Bacterial Communities in Human Neonates". Applied and Environmental Microbiology. 68 (1): 219–26. doi:10.1128/AEM.68.1.219-226.2002. PMC 126580. PMID 11772630.
- ^ Coppa, Giovanni V; Bruni, Stefano; Morelli, Lorenzo; Soldi, Sara; Gabrielli, Orazio (2004). "The First Prebiotics in Humans". Journal of Clinical Gastroenterology. 38 (6 Suppl): S80–3. doi:10.1097/01.mcg.0000128926.14285.25. PMID 15220665.
- ^ Coppa, G.V.; Zampini, L.; Galeazzi, T.; Gabrielli, O. (2006). "Prebiotics in human milk: A review". Digestive and Liver Disease. 38: S291–4. doi:10.1016/S1590-8658(07)60013-9. PMID 17259094.
- ^ Harmsen, Hermie J. M.; Wildeboer-Veloo, Alida C. M.; Raangs, Gerwin C.; Wagendorp, Arjen A.; Klijn, Nicolette; Bindels, Jacques G.; Welling, Gjalt W. (2000). "Analysis of Intestinal Flora Development in Breast-Fed and Formula-Fed Infants by Using Molecular Identification and Detection Methods". Journal of Pediatric Gastroenterology and Nutrition. 30 (1): 61–7. doi:10.1097/00005176-200001000-00019. PMID 10630441.
- ^ Fanaro, S; Chierici, R; Guerrini, P; Vigi, V (2003). "Intestinal microflora in early infancy: Composition and development". Acta Paediatrica. 91 (441): 48–55. PMID 14599042.
- ^ "Discordant temporal development of bacterial phyla and the emergence of core in the fecal microbiota of young children". The ISME Journal. 2015.
- ^ a b Gerritsen, Jacoline; Smidt, Hauke; Rijkers, Ger; de Vos, Willem (27 May 2011). "Intestinal microbiota in human health and disease: the impact of probiotics". Genes & Nutritions. 6 (3): 209–240. doi:10.1007/s12263-011-0229-7. PMC 3145058. PMID 21617937.
- ^ a b c d e f g Yatsunenko, T.; Rey, F. E.; Manary, M. J.; Trehan, I.; Dominguez-Bello, M. G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R. N.; Anokhin, A. P.; Heath, A. C.; Warner, B.; Reeder, J.; Kuczynski, J.; Caporaso, J. G.; Lozupone, C. A.; Lauber, C.; Clemente, J. C.; Knights, D.; Knight, R.; Gordon, J. I. (2012). "Human gut microbiome viewed across age and geography". Nature. 486 (7402): 222–227. Bibcode:2012Natur.486..222Y. doi:10.1038/nature11053. PMC 3376388. PMID 22699611. Cite error: The named reference "Tanya Yatsunenko 2012" was defined multiple times with different content (see the help page).
- ^ "Keen to be healthier in old age? Tend your inner garden". The conversation. 2016.
- ^ Jackson, Matthew A; Jeffery, Ian B; Beaumont, Michelle; Bell, Jordana T; Clark, Andrew G; Ley, Ruth E; o'Toole, Paul W; Spector, Tim D; Steves, Claire J (2016). "Signatures of early frailty in the gut microbiota". Bio Med Central. 8. doi:10.1186/s13073-016-0262-7.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Jonkers, Daisy M.A.E. (2016). "Microbial perturbations and modulation in conditions associated with malnutrition and malabsorption". Best Practice & Research Clinical Gastroenterology. 30 (2): 161–172. doi:10.1016/j.bpg.2016.02.006. PMID 27086883.
- ^ Million, Matthieu; Diallo, Aldiouma; Raoult, Didier (2016). "Gut microbiota and malnutrition" (PDF). Microbial Pathogenesis. 106: 127–138. doi:10.1016/j.micpath.2016.02.003. PMID 26853753.
- ^ Rytter, Maren Johanne Heilskov; Kolte, Lilian (2014). "The Immune System in Children with Malnutrition". PLoS. 9 (8): e105017. Bibcode:2014PLoSO...9j5017R. doi:10.1371/journal.pone.0105017. PMC 4143239. PMID 25153531.
{{cite journal}}: Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help)CS1 maint: unflagged free DOI (link) - ^ Cite error: The named reference
:0was invoked but never defined (see the help page). - ^ a b "Western diets damage gut microbiota over generations, in ways hard to reverse". La Times. 2016.
- ^ "Gut microbiota need fiber, too". The-scientist.com. 2016.
- ^ "Probiotics: In Depth". National Center for Complementary and Integrative Health. October 2016.
- ^ De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J. B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. (2010). "Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa". Proc. Natl. Acad. Sci. U.S.A. 107 (33): 14691–14696. Bibcode:2010PNAS..10714691D. doi:10.1073/pnas.1005963107. PMC 2930426. PMID 20679230.
- ^ "Early-life exercise may promote lasting brain and metabolic health through gut bacterial metabolites". Immunology and Cell Biology. 2016.
- ^ David, Lawrence A; Materna, Arne C; Friedman, Jonathan; Campos-Baptista, Maria I; Blackburn, Matthew C; Perrotta, Allison; Erdman, Susan E; Alm, Eric J (2014). "Host lifestyle affects human microbiota on daily timescales". Genome Biology. 15 (7): R89. doi:10.1186/gb-2014-15-7-r89.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Keeley J. 2004. Good bacteria trigger proteins to protect the gut. Howard Hughes Medical Institute. EurekAlert. Accessed January 9, 2007
- ^ a b Wynne, Anthony G; McCartney, Anne L; Brostoff, Jonathan; Hudspith, Barry N; Gibson, Glenn R (2004). "An in vitro assessment of the effects of broad-spectrum antibiotics on the human gut microflora and concomitant isolation of a Lactobacillus plantarum with anti-Candida activities". Anaerobe. 10 (3): 165–9. doi:10.1016/j.anaerobe.2004.03.002. PMID 16701514.
- ^ a b c d e f "Can the bacteria in your gut explain your mood?". New York Times. 2015.
- ^ http://www.pnas.org/content/28/7/285?ijkey=931998456351a78f5d633e70cbc46b08369e0414&keytype2=tf_ipsecsha
- ^ https://www.clinicalkey.com/#!/ContentPlayerCtrl/doPlayContent/1-s2.0-S095816691200119X
- ^ Cummings, J.H.; MacFarlane, G.T. (1997). "Role of intestinal bacteria in nutrient metabolism". Clinical Nutrition. 16: 3–9. doi:10.1016/S0261-5614(97)80252-X.
- ^ a b c d e Björkstén, Bengt; Sepp, Epp; Julge, Kaja; Voor, Tiia; Mikelsaar, Marika (2001). "Allergy development and the intestinal microflora during the first year of life". Journal of Allergy and Clinical Immunology. 108 (4): 516–20. doi:10.1067/mai.2001.118130. PMID 11590374.
- ^ Savage, D C (1977). "Microbial Ecology of the Gastrointestinal Tract". Annual Review of Microbiology. 31: 107–33. doi:10.1146/annurev.mi.31.100177.000543. PMID 334036.
- ^ Adam, Clare L; Williams, Patricia A; Dalby, Matthew J; Garden, Karen; Thomson, Lynn M; Richardson, Anthony J; Gratz, Silvia W; Ross, Alexander W (2014). "Different types of soluble fermentable dietary fibre decrease food intake, body weight gain and adiposity in young adult male rats". Nutrition and metabolism. 11: 36. doi:10.1186/1743-7075-11-36.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ "The role of short chain fatty acids in appetite regulation and energy homeostasis". International Journal of Obesity. 2015.
- ^ Hayward, A. R (1983). "The human fetus and newborn: development of the immune response". Birth Defects Original Article Series. 19 (3): 289–94. PMID 6606446.
- ^ a b c d e f g h i j Cite error: The named reference
Sommer2013revwas invoked but never defined (see the help page). - ^ Cite error: The named reference
Faderl2015revwas invoked but never defined (see the help page). - ^ Reinoso Webb C (2016). "Protective and pro-inflammatory roles of intestinal bacteria". Pathophysiology (Review). 23 (2): 67–80. doi:10.1016/j.pathophys.2016.02.002. PMC 4867289. PMID 26947707.
- ^ Mantis NJ, Rol N, Corthésy B (2011). "Secretory IgA's complex roles in immunity and mucosal homeostasis in the gut". Mucosal Immunol. 4 (6): 603–11. doi:10.1038/mi.2011.41. PMC 3774538. PMID 21975936.
- ^ "Battle in the gut: Immune cells help 'good bacteria' triumph over 'bad bacteria'". Cell Press. 2015.
- ^ Jewell, A.P. (2005). "Is the liver an important site for the development of immune tolerance to tumours?". Medical Hypotheses. 64 (4): 751–4. doi:10.1016/j.mehy.2004.10.002. PMID 15694692.
- ^ Tlaskalová-Hogenová H., Stepánková R., Hudcovic T., Tucková L., Cukrowska B., Lodinová-Zádníková R., Kozáková H., Rossmann P., Bártová J., Sokol D.; et al. (2004). "Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases". Immunol. Lett. 93 (2–3): 97–108. doi:10.1016/j.imlet.2004.02.005. PMID 15158604.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Penders J., Stobberingh E. E., van den Brandt P. A., Thijs C. (2007). "The role of the intestinal microbiota in the development of atopic disorders". Allergy. 62 (11): 1223–36. doi:10.1111/j.1398-9995.2007.01462.x. PMID 17711557.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Björkstén B., Sepp E., Julge K., Voor T., Mikelsaar M. (2001). "Allergy development and the intestinal microflora during the first year of life". J. Allergy Clin. Immunol. 108 (4): 516–20. doi:10.1067/mai.2001.118130. PMID 11590374.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ "'Hygiene Hypothesis' or 'Old Friends' Theory?". The Huffington Post. 2012.
- ^ Sheikh, A; Strachan, D. P (2004). "The hygiene theory: fact or fiction?". Current Opinion in Otolaryngology & Head and Neck Surgery. 12 (3): 232–6. PMID 15167035.
- ^ Okada, H; Kuhn, C; Feillet, H; Bach, J. F (2010). "The 'hygiene hypothesis' for autoimmune and allergic diseases: an update". Clinical & Experimental Immunology. 160 (1): 1–9. doi:10.1111/j.1365-2249.2010.04139.x. PMC 2841828. PMID 20415844.
- ^ a b c "Gut microbiota regulates antioxidant metabolism". Science Daily. 2015.
- ^ a b c d e f g "Gut microbiota and brain function: An evolving field in neuroscience" (PDF). CINP. 2015.
- ^ a b c d e f g h i Zhang LS, Davies SS (April 2016). "Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions". Genome Med. 8 (1): 46. doi:10.1186/s13073-016-0296-x. PMC 4840492. PMID 27102537.
Lactobacillus spp. convert tryptophan to indole-3-aldehyde (I3A) through unidentified enzymes [125]. Clostridium sporogenes convert tryptophan to IPA [6], likely via a tryptophan deaminase. ... IPA also potently scavenges hydroxyl radicals
Table 2: Microbial metabolites: their synthesis, mechanisms of action, and effects on health and disease
Figure 1: Molecular mechanisms of action of indole and its metabolites on host physiology and disease - ^ Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G (March 2009). "Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites". Proc. Natl. Acad. Sci. U.S.A. 106 (10): 3698–3703. Bibcode:2009PNAS..106.3698W. doi:10.1073/pnas.0812874106. PMC 2656143. PMID 19234110.
Production of IPA was shown to be completely dependent on the presence of gut microflora and could be established by colonization with the bacterium Clostridium sporogenes.
IPA metabolism diagram - ^ "3-Indolepropionic acid". Human Metabolome Database. University of Alberta. Retrieved 12 June 2018.
- ^ Chyan YJ, Poeggeler B, Omar RA, Chain DG, Frangione B, Ghiso J, Pappolla MA (July 1999). "Potent neuroprotective properties against the Alzheimer beta-amyloid by an endogenous melatonin-related indole structure, indole-3-propionic acid". J. Biol. Chem. 274 (31): 21937–21942. doi:10.1074/jbc.274.31.21937. PMID 10419516. S2CID 6630247.
[Indole-3-propionic acid (IPA)] has previously been identified in the plasma and cerebrospinal fluid of humans, but its functions are not known. ... In kinetic competition experiments using free radical-trapping agents, the capacity of IPA to scavenge hydroxyl radicals exceeded that of melatonin, an indoleamine considered to be the most potent naturally occurring scavenger of free radicals. In contrast with other antioxidants, IPA was not converted to reactive intermediates with pro-oxidant activity.
- ^ a b c d e Cite error: The named reference
2014Wangrevwas invoked but never defined (see the help page). - ^ Mayer EA, Knight R, Mazmanian SK (2014). "Gut microbes and the brain: paradigm shift in neuroscience" (PDF). J Neurosci. 34 (46): 15490–15496. doi:10.1523/JNEUROSCI.3299-14.2014. PMC 4228144. PMID 25392516.
{{cite journal}}: Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help) - ^ a b c Dinan, T.G; Cryan, 2015 (2015). "The impact of gut microbiota on brain and behavior: implications for psychiatry". Curr Opin Clin Nutr Metab Care. 18 (6): 552–558. doi:10.1097/MCO.0000000000000221. PMID 26372511.
{{cite journal}}:|first2=has numeric name (help) - ^ a b c Schneiderhan J, Master-Hunter T, Locke A (2016). "Targeting gut flora to treat and prevent disease". J Fam Pract. 65 (1): 34–8. PMID 26845162.
- ^ Wang H, Lee IS, Braun C, Enck P (July 2016). "Effect of probiotics on central nervous system functions in animals and humans - a systematic review". J. Neurogastroenterol Motil. 22 (4): 589–605. doi:10.5056/jnm16018. PMC 5056568. PMID 27413138.
We reviewed the effect of probiotics on the central nervous system in randomized controlled trials in animals and humans, and analyzed the possibility of translating animal models to human studies because few human studies have been conducted to date. According to the qualitative analyses of current studies, we can provisionally draw the conclusion that B. longum, B. breve, B. infantis, L. helveticus, L. rhamnosus, L. plantarum, and L. casei were most effective in improving CNS function, including psychiatric disease-associated functions (anxiety, depression, mood, stress response) and memory abilities.
- ^ "Antibiotics and the gut microbiome". The-scientist.com. 2015.
- ^ a b c d e f Cho, I.; Yamanishi, S.; Cox, L.; Methé, B. A.; Zavadil, J.; Li, K.; Gao, Z.; Mahana, D.; Raju, K.; Teitler, I.; Li, H.; Alekseyenko, A. V.; Blaser, M. J. (2012). "Antibiotics in early life alter the murine colonic microbiome and adiposity". Nature. 488 (7413): 621–6. Bibcode:2012Natur.488..621C. doi:10.1038/nature11400. PMC 3553221. PMID 22914093.
- ^ a b Carman, Robert J.; Simon, Mary Alice; Fernández, Haydée; Miller, Margaret A.; Bartholomew, Mary J. (2004). "Ciprofloxacin at low levels disrupts colonization resistance of human fecal microflora growing in chemostats". Regulatory Toxicology and Pharmacology. 40 (3): 319–26. doi:10.1016/j.yrtph.2004.08.005. PMID 15546686.
- ^ "Some antibiotics may change gut bacteria in kids". Fox News. 2016.
- ^ "The composition of the gut microbiota throughout life, with an emphasis on early life". Microbial Ecology in health and disease. 2016.
- ^ a b Knight, DJW; Girling, KJ (2003). "Gut flora in health and disease". The Lancet. 361 (9371): 512–9. doi:10.1016/S0140-6736(03)13438-1. PMID 12781578.
- ^ "Food Additives Linked to Inflammation". The-scientist.com. 2015.
- ^ a b "Microbes play role in anti tumor response". The-scientist.com. 2015.
- ^ a b c d Guarner, Francisco; Malagelada, Juan-R (2003). "Role of bacteria in experimental colitis". Best Practice & Research Clinical Gastroenterology. 17 (5): 793–804. doi:10.1016/S1521-6918(03)00068-4. PMID 14507589.
- ^ a b Hugot, Jean-Pierre (2004). "Inflammatory bowel disease: A complex group of genetic disorders". Best Practice & Research Clinical Gastroenterology. 18 (3): 451–62. doi:10.1016/j.bpg.2004.01.001. PMID 15157820.
- ^ Veltkamp, Claudia; Tonkonogy, Susan L.; De Jong, Ype P.; Albright, Carol; Grenther, Wetonia B.; Balish, Edward; Terhorst, Cox; Sartor, R.Balfour (2001). "Continuous stimulation by normal luminal bacteria is essential for the development and perpetuation of colitis in Tgϵ26 mice". Gastroenterology. 120 (4): 900–13. doi:10.1053/gast.2001.22547. PMID 11231944.
- ^ "Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn's Disease". American Society for Microbiology. 2016.
- ^ a b c Suenaert, Peter; Bulteel, Veerle; Lemmens, Liesbeth; Noman, Maja; Geypens, Benny; Van Assche, Gert Van; Geboes, Karel; Ceuppens, Jan L.; Rutgeerts, Paul (2002). "Anti-tumor necrosis factor treatment restores the gut barrier in Crohn's disease". The American Journal of Gastroenterology. 97 (8): 2000–4. doi:10.1111/j.1572-0241.2002.05914.x. PMID 12190167.
- ^ a b c d e "Probiotics Pros and Cons". Berkeley Wellness. 2014.
- ^ Cite error: The named reference
Shen2016revwas invoked but never defined (see the help page). - ^ Burisch, Johan; Jess, Tine; Martinato, Matteo; Lakatos, Peter L. (2013). "The burden of inflammatory bowel disease in Europe". Journal of Crohn's and Colitis. 7 (4): 322–337. doi:10.1016/j.crohns.2013.01.010. ISSN 1873-9946. PMID 23395397.
- ^ Blandino G (2016). "Impact of gut microbiota on diabetes mellitus". Diabetes Metab. (Review). 42 (5): 303–315. doi:10.1016/j.diabet.2016.04.004. PMID 27179626.
{{cite journal}}: Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help) pii: S1262-3636(16)30396-2 - ^ Boulangé CL (2016). "Impact of the gut microbiota on inflammation, obesity, and metabolic disease". Genome Med. (Review). 8 (1): 42. doi:10.1186/s13073-016-0303-2. PMC 4839080. PMID 27098727.
{{cite journal}}: Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help)CS1 maint: unflagged free DOI (link) - ^ a b Cite error: The named reference
Saxena2016was invoked but never defined (see the help page). - ^ "What are the consequences of the disappearing human microbiota?". Nature Reviews Microbiology. 2009.
- ^ "Does this phylum make me look fat". NPR. 2015.
- ^ Wendelsdorf, Katherine. (2013). "Gut microbiota from twins discordant for obesity modulate metabolism in mice". Science. 341 (6150): 1241214. doi:10.1126/science.1241214. PMC 3829625. PMID 24009397.
- ^ Ley, Ruth E.; Turnbaugh, Peter J.; Klein, Samuel; Gordon, Jeffrey I. (2006). "Microbial ecology: Human gut microbes associated with obesity". Nature. 444 (7122): 1022–3. doi:10.1038/4441022a. PMID 17183309.
- ^ Turnbaugh, Peter J.; Ley, Ruth E.; Mahowald, Michael A.; Magrini, Vincent; Mardis, Elaine R.; Gordon, Jeffrey I. (2006). "An obesity-associated gut microbiome with increased capacity for energy harvest". Nature. 444 (7122): 1027–31. doi:10.1038/nature05414. PMID 17183312.
- ^ Backhed, F.; Manchester, J. K.; Semenkovich, C. F.; Gordon, J. I. (2007). "Mechanisms underlying the resistance to diet-induced obesity in germ-free mice". Proceedings of the National Academy of Sciences. 104 (3): 979–984. doi:10.1073/pnas.0605374104. PMC 1764762. PMID 17210919.
- ^ Backhed, F.; Manchester, JK; Semenkovich, CF; Gordon, JI (2004). "The gut microbiota as an environmental factor that regulates fat storage". Proceedings of the National Academy of Sciences. 101 (44): 979–84. doi:10.1073/pnas.0407076101. PMC 1764762. PMID 17210919.
- ^ Walker, A. W.; Parkhill, J. (2013). "Fighting Obesity with Bacteria". Scrience. 341 (6150): 1069–1070. doi:10.1126/science.1243787.
- ^ "Microbesity". The-scientist.com. 2015.
- ^ "Inside the connection between bacterial toxins and obesity". Digital Journal. 2016.
- ^ "The importance of the gut microbiota after bariatric surgery". Nature. 2012.
- ^ a b Murri, Mora; Leiva, Isabel; Gomez-Zumaquero, Juan Miguel; Tinahones, Francisco J; Cardona, Fernando; Soriguer, Federico; Queipo-Ortuño, María Isabel (2013). "Gut microbiota in children with type 1 diabetes differs from that in healthy children". BMC Medicine. 11. doi:10.1186/1741-7015-11-46.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ "Can gut bacteria be used for early diagnosis of type 2 diabetes?". Gut microbiota for health. 2016.
- ^ "The key to your brain's health lies in your gut, says neurologist". Stuff. 2016.
- ^ a b c d "The bacteria in our guts may help determine who gets anxiety and depression". Science News. 2016.
- ^ Carabotti, M; Scirocco, A; Maselli, M. A; Severi, C (2015). "The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems". Annals of Gastroenterology. 28 (2): 203–209. PMC 4367209. PMID 25830558.
- ^ "9 ways your microbiome influences your weight and health". Mindbodygreen.com. 2016.
- ^ "Gut bacteria may help prevent stroke:study". Tech Times. 2016.
- ^ a b "Changing gut bacteria through diet affects brain function, UCLA study shows". UCLA. 2013.
- ^ a b "Why a psychiatrist thinks the key to happiness might be swallowing the right bacteria". Business Insider. 2016.
- ^ "Protein from bacteria alleviates food allergy symptoms". Institute for Basic Science. 2016.
- ^ "Allergies and the Benefits of Yogurt". Sharecare. 2010.
- ^ "Microbiome: Gut reaction". Nature. 2011.
- ^ a b Minemura M, Shimizu Y (2015). "Gut microbiota and liver diseases". World J Gastroenterol. 21 (6): 1691–702. doi:10.3748/wjg.v21.i6.1691. PMC 4323444. PMID 25684933.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ "gut microbiota alterations can predict hospitalizations in cirrhosis". Nature. 2015.
- ^ Garcia-Tsao, Guadalupe; Wiest, Reiner (2004). "Gut microflora in the pathogenesis of the complications of cirrhosis". Best Practice & Research Clinical Gastroenterology. 18 (2): 353–72. doi:10.1016/j.bpg.2003.10.005. PMID 15123075.
- ^ "Visceral Pain and Gastrointestinal Microbiome". Journal of Neurogastroenterology and Motility. 2015.
- ^ "Probiotic Guidelines" (PDF). World Health Organization. 2002.
- ^ Ford, Alexander C; Quigley, Eamonn M M; Lacy, Brian E; Lembo, Anthony J; Saito, Yuri A; Schiller, Lawrence R; Soffer, Edy E; Spiegel, Brennan M R; Moayyedi, Paul (2014). "Efficacy of Prebiotics, Probiotics, and Synbiotics in Irritable Bowel Syndrome and Chronic Idiopathic Constipation: Systematic Review and Meta-analysis". The American Journal of Gastroenterology. 109 (10): 1547–1561. doi:10.1038/ajg.2014.202. ISSN 0002-9270. PMID 25070051.
- ^ Ghouri, Yezaz A; Richards, David M; Rahimi, Erik F; Krill, Joseph T; Jelinek, Katherine A; DuPont, Andrew W (2014). "Systematic review of randomized controlled trials of probiotics, prebiotics, and synbiotics in inflammatory bowel disease". Clin Exp Gastroenterol. 7: 473–487. doi:10.2147/CEG.S27530. PMC 4266241. PMID 25525379.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Yu CG, Huang Q (2013). "Recent progress on the role of gut microbiota in the pathogenesis of inflammatory bowel disease". J Dig Dis. 14 (10): 513–7. doi:10.1111/1751-2980.12087. PMID 23848393.
- ^ Hickson, M (2011). "Probiotics in the prevention of antibiotic-associated diarrhoea and Clostridium difficile infection". Therapeutic Advances in Gastroenterology. 4 (3): 185–197. doi:10.1177/1756283X11399115. PMC 3105609. PMID 21694803.
- ^ Kim, Eun-Hee; Hong, Hua; Choi, Ki-Seok; Han, Young-Min; Kangwan, Napapan; Cho, Young Chae; Hahm, Ki Baik (April 2012). "High Concentrated Probiotics Improve Inflammatory Bowel Diseases Better than Commercial Concentration of Probiotics". Journal of Food & Drug Analysis. 20: 292–5.
- ^ Hungin, A. P; Mulligan, C; Pot, B; Whorwell, P; Agréus, L; Fracasso, P; Lionis, C; Mendive, J; Philippart De Foy, J. M; Rubin, G; Winchester, C; Wit, N; European Society for Primary Care Gastroenterology (2013). "Systematic review: probiotics in the management of lower gastrointestinal symptoms in clinical practice – an evidence-based international guide". Alimentary Pharmacology & Therapeutics. 38 (8): 864–886. doi:10.1111/apt.12460. PMC 3925990. PMID 23981066.
- ^ "Gastrointestinal effects of prebiotics". British Journal of Nutrition. 2002.
- ^ Min, Y. N; Yang, H. L; Xu, Y. X; Gao, Y. P (2016). "Effects of dietary supplementation of synbiotics on growth performance, intestinal morphology, sIgA content and antioxidant capacities of broilers". Journal of animal physiology and animal nutrition. 100 (6): 1073–1080. doi:10.1111/jpn.12479.
- ^ Engel, P.; Moran, N. (2013). "The gut microbiota of insects–diversity in structure and function". FEMS Microbiology Reviews. 5 (5): 699–735. doi:10.1111/1574-6976.12025.
- ^ Brune, A. (2014). "Symbiotic digestion of lignocellulose in termite guts". Nature Review Microbiology. 12 (3): 168–180. doi:10.1038/nrmicro3182.
- ^ a b Dietrich, C.; Köhler, T.; Brune, A. (2014). "The cockroach origin of the termite gut microbiota: patterns in bacterial community structure reflect major evolutionary events". Applied and Environmental Microbiology. 80 (7): 2261–2269. doi:10.1128/AEM.04206-13. PMC 3993134. PMID 24487532.
- ^ a b Mikaelyan, A.; Dietrich, C.; Köhler, T.; Poulsen, M.; Sillam-Dussès, D.; Brune, A. (2015). "Diet is the primary determinant of bacterial community structure in the guts of higher termites". Molecular Ecology. 24 (20): 5824–5895. doi:10.1111/mec.13376. PMID 26348261.
- ^ Mikaelyan, A.; Thompson, C.; Hofer, M.; Brune, A. (2016). "The deterministic assembly of complex bacterial communities in germ-free cockroach guts". Applied and Environmental Microbiology. 82 (4): 1256–1263. doi:10.1128/AEM.03700-15. PMID 26655763.
Further reading[edit]
- Review articles
- Maranduba, CM; De Castro, SB; de Souza, GT; Rossato, C; da Guia, FC; Valente, MA; Rettore, JV; Maranduba, CP; de Souza, CM; do Carmo, AM; Macedo, GC; Silva, FS (2015). "Intestinal Microbiota as Modulators of the Immune System and Neuroimmune System: Impact on the Host Health and Homeostasis". Journal of Immunology Research. 2015: 1–14. doi:10.1155/2015/931574. PMC 4352473. PMID 25759850.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - De Preter, Vicky; Hamer, Henrike M; Windey, Karen; Verbeke, Kristin (2011). "The impact of pre- and/or probiotics on human colonic metabolism: Does it affect human health?". Molecular Nutrition & Food Research. 55 (1): 46–57. doi:10.1002/mnfr.201000451. PMID 21207512.
- Prakash, Satya; Rodes, Laetitia; Coussa-Charley, Michael; Tomaro-Duchesneau, Catherine; Tomaro-Duchesneau, Catherine; Coussa-Charley; Rodes (2011). "Gut microbiota: Next frontier in understanding human health and development of biotherapeutics". Biologics: Targets and Therapy. 5: 71–86. doi:10.2147/BTT.S19099. PMC 3156250. PMID 21847343.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - Wu, G. D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S. A.; Bewtra, M.; Knights, D.; Walters, W. A.; Knight, R.; Sinha, R.; Gilroy, E.; Gupta, K.; Baldassano, R.; Nessel, L.; Li, H.; Bushman, F. D.; Lewis, J. D. (2011). "Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes". Science. 334 (6052): 105–8. Bibcode:2011Sci...334..105W. doi:10.1126/science.1208344. PMC 3368382. PMID 21885731.