User:Awchen/sandbox
| Elongation Factor Thermo Unstable | |||||||||
|---|---|---|---|---|---|---|---|---|---|
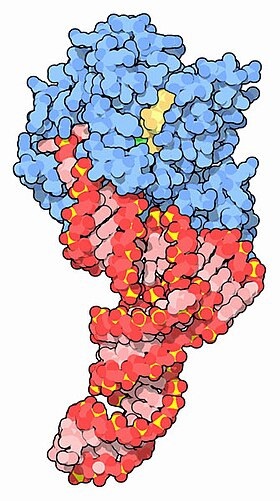 EF-Tu (blue) complexed with tRNA (red) and GTP (yellow) [1] | |||||||||
| Identifiers | |||||||||
| Symbol | EF-Tu | ||||||||
| Pfam | GTP_EFTU | ||||||||
| InterPro | IPR004541 | ||||||||
| CATH | 1ETU | ||||||||
| SCOP2 | 1ETU / SCOPe / SUPFAM | ||||||||
| |||||||||
EF-Tu (elongation factor thermo unstable) is a prokaryotic elongation factor responsible for catalyzing the binding of an aminoacyl-tRNA (aa-tRNA) to the ribosome. It is a G-protein, and facilitates the selection and binding of an aa-tRNA to the A-site of the ribosome. As a reflection of its crucial role in translation, EF-Tu is one of the most abundant and highly conserved proteins in prokaryotes.[2][3][4]
Background[edit]
Elongation factors are part of the mechanism that synthesizes new proteins through translation in the ribosome. Transfer RNAs (tRNAs) carry the individual amino acids that become integrated into a protein sequence, and have an anticodon for the specific amino acid that they are charged with. Messenger RNA (mRNA) carries the genetic information that encodes the primary structure of a protein, and contains codons that code for each amino acid. The ribosome creates the protein chain by following the mRNA code and integrating the amino acid of an aminoacyl-tRNA (also known as a charged tRNA) to the growing polypeptide chain.[5][6]
There are three sites on the ribosome for tRNA binding. These are the aminoacyl/acceptor site (abbreviated A), the peptidyl site (abbreviated P), and the exit site (abbreviated E). The P-site holds the tRNA connected to the polypeptide chain being synthesized, and the A-site is the binding site for a charged tRNA with an anticodon complementary to the mRNA codon associated with the site. After binding of a charged tRNA to the A-site, a peptide bond is formed between the growing polypeptide chain on the P-site tRNA and the amino acid of the A-site tRNA, and the entire polypeptide is transferred from the P-site tRNA to the A-site tRNA. Then, in a process catalyzed by the prokaryotic elongation factor EF-G (historically known as translocase), the coordinated translocation of the tRNAs and mRNA occurs, with the P-site tRNA moving to the E-site, where it dissociates from the ribosome, and the A-site tRNA moves to take its place in the P-site.[5][6]
Biological Functions[edit]

Protein Synthesis[edit]
EF-Tu participates in the polypeptide elongation process of protein synthesis. In prokaryotes, the primary function of EF-Tu is to transport the correct aa-tRNA to the A-site of the ribosome. As a G-protein, it uses GTP to facilitate its function. Outside of the ribosome, EF-Tu complexed with GTP (EF-Tu • GTP) complexes with aa-tRNA to form a stable EF-Tu • GTP • aa-tRNA ternary complex.[7] EF-Tu • GTP binds all correctly-charged aa-tRNAs with approximately identical affinity, except those charged with initiation residues and selenocysteine.[8][9] This can be accomplished because although different amino acid residues have varying side-chain properties, the tRNAs associated with those residues have varying structures to compensate for differences in side-chain binding affinities.[10][11]
The binding of an aa-tRNA to EF-Tu • GTP allows for the ternary complex to be translocated to the A-site of an active ribosome, in which the anticodon of the tRNA binds to the codon of the mRNA. If the correct anticodon binds to the mRNA codon, the ribosome changes configuration and alters the geometry of the GTPase domain of EF-Tu, resulting in the hydrolysis of the GTP associated with the EF-Tu to GDP and Pi. As such, the ribosome functions as a GTPase-activating protein (GAP) for EF-Tu. Upon GTP hydrolysis, the conformation of EF-Tu changes drastically and dissociates from the aa-tRNA and ribosome complex.[4][12] The aa-tRNA then fully enters the A-site, where its amino acid is brought near the P-site's polypeptide and the ribosome catalyzes the covalent transfer of the polypeptide onto the amino acid.[9]
In the cytoplasm, the deactivated EF-Tu • GDP is acted on by the prokaryotic elongation factor EF-Ts, which causes EF-Tu to release its bound GDP. Upon dissociation of EF-Ts, EF-Tu is able to complex with a GTP due to the 5– to 10–fold higher concentration of GTP than GDP in the cytoplasm, resulting in reactivated EF-Tu • GTP, which can then associate with another aa-tRNA.[7][12]
Maintaining Translational Accuracy[edit]
EF-Tu contributes to translational accuracy in three ways. In translation, a fundamental problem is that near-cognate anticodons have similar binding affinity to a codon as cognate anticodons, such that anticodon-codon binding in the ribosome alone is not sufficient to maintain high translational fidelity. This is addressed by the ribosome not activating the GTPase activity of EF-Tu if the tRNA in the ribosome's A-site does not match the mRNA codon, thus preferentially increasing the likelihood for the incorrect tRNA to leave the ribosome.[13] Additionally, regardless of tRNA matching, EF-Tu also induces a delay after freeing itself from the aa-tRNA, before the aa-tRNA fully enters the A-site (a process called accommodation). This delay period is a second opportunity for incorrectly charged aa-tRNAs to move out of the A-site before the incorrect amino acid is irreversibly added to the polypeptide chain.[14][15] A third mechanism is the less well understood function of EF-Tu to crudely check aa-tRNA associations and reject complexes where the amino acid is not bound to the correct tRNA coding for it.[10]
Other Functions[edit]
EF-Tu has been found in large quantities in the cytoskeletons of bacteria, co-localizing underneath the cell membrane with MreB, a cytoskeletal element that maintains cell shape.[16][17] Defects in EF-Tu have been shown to result in defects in bacterial morphology.[18] Additionally, EF-Tu has displayed some chaperone-like characteristics, with some experimental evidence suggesting that it promotes the refolding of a number of denatured proteins in vitro.[19][20]
Structure[edit]

EF-Tu is a monomeric protein with molecular weight around 43 kDa in Escherichia coli.[21][22][23] The protein consists of three structural domains: a GTP-binding domain and two oligonucleotide-binding domains. The N-terminal domain I of EF-Tu is the GTP-binding domain. It consists of a six beta-strand core flanked by six alpha-helices.[7] Domains II and III of EF-Tu, the oligonucleotide-binding domains, both adopt beta-barrel structures.[24][25]
The GTP-binding domain I undergoes a dramatic conformational change upon GTP hydrolysis to GDP, allowing EF-Tu to dissociate from aa-tRNA and leave the ribosome.[26] Reactivation of EF-Tu is achieved by GTP binding in the cytoplasm, which leads to a significant conformational change that reactivates the tRNA-binding site of EF-Tu. In particular, GTP binding to EF-Tu results in a ~90° rotation of domain I relative to domains II and III, exposing the residues of the tRNA-binding active site.[27]
Disease Relevance[edit]
Along with the ribosome, EF-Tu is one of the most important targets for antibiotic-mediated inhibition of translation.[7] Antibiotics targeting EF-Tu can be categorized into one of two groups, depending on the mechanism of action, and one of four structural families. The first group includes the antibiotics pulvomycin and GE2270A, and inhibits the formation of the ternary complex.[28] The second group includes the antibiotics kirromycin and enacyloxin, and prevents the release of EF-Tu from the ribosome after GTP hydrolysis.[29][30][31]
See also[edit]
References[edit]
- ^ PDB Molecule of the Month EF-Tu
- ^ Weijland, A.; Harmark, K.; Cool, R. H.; Anborgh, P. H.; Parmeggiani, A. (1992-03-01). "Elongation factor Tu: a molecular switch in protein biosynthesis". Molecular Microbiology. 6 (6): 683–688. doi:10.1111/j.1365-2958.1992.tb01516.x. ISSN 0950-382X. PMID 1573997. S2CID 10966743.
- ^ "TIGR00485: EF-Tu". National Center for Biotechnology Information. March 3, 2017.
- ^ a b Yamamoto, Hiroshi; Qin, Yan; Achenbach, John; Li, Chengmin; Kijek, Jaroslaw; Spahn, Christian M. T.; Nierhaus, Knud H. (2014-02-01). "EF-G and EF4: translocation and back-translocation on the bacterial ribosome". Nature Reviews. Microbiology. 12 (2): 89–100. doi:10.1038/nrmicro3176. ISSN 1740-1534. PMID 24362468. S2CID 27196901.
- ^ a b Laursen, Brian Søgaard; Sørensen, Hans Peter; Mortensen, Kim Kusk; Sperling-Petersen, Hans Uffe (2005-03-01). "Initiation of protein synthesis in bacteria". Microbiology and Molecular Biology Reviews: MMBR. 69 (1): 101–123. doi:10.1128/MMBR.69.1.101-123.2005. ISSN 1092-2172. PMC 1082788. PMID 15755955.
- ^ a b Ramakrishnan, V. (2002-02-22). "Ribosome structure and the mechanism of translation". Cell. 108 (4): 557–572. doi:10.1016/s0092-8674(02)00619-0. ISSN 0092-8674. PMID 11909526. S2CID 2078757.
- ^ a b c d Krab, Ivo M.; Parmeggiani, Andrea (2002-01-01). "Mechanisms of EF-Tu, a pioneer GTPase". Progress in Nucleic Acid Research and Molecular Biology. 71: 513–551. doi:10.1016/s0079-6603(02)71050-7. ISBN 9780125400718. ISSN 0079-6603. PMID 12102560.
- ^ "Translation elongation factor EFTu/EF1A, bacterial/organelle (IPR004541)". InterPro.
- ^ a b Diwan, Joyce (2008). "Translation: Protein Synthesis". Rensselaer Polytechnic Institute.
- ^ a b LaRiviere, F. J.; Wolfson, A. D.; Uhlenbeck, O. C. (2001-10-05). "Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation". Science (New York, N.Y.). 294 (5540): 165–168. doi:10.1126/science.1064242. ISSN 0036-8075. PMID 11588263. S2CID 26192336.
- ^ Louie, A.; Ribeiro, N. S.; Reid, B. R.; Jurnak, F. (1984-04-25). "Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP". The Journal of Biological Chemistry. 259 (8): 5010–5016. doi:10.1016/S0021-9258(17)42947-4. ISSN 0021-9258. PMID 6370998.
- ^ a b Clark, B. F.; Nyborg, J. (1997-02-01). "The ternary complex of EF-Tu and its role in protein biosynthesis". Current Opinion in Structural Biology. 7 (1): 110–116. doi:10.1016/s0959-440x(97)80014-0. ISSN 0959-440X. PMID 9032056.
- ^ Nilsson, Jakob; Nissen, Poul (2005-06-01). "Elongation factors on the ribosome". Current Opinion in Structural Biology. 15 (3): 349–354. doi:10.1016/j.sbi.2005.05.004. ISSN 0959-440X. PMID 15922593.
- ^ Whitford, Paul C.; Geggier, Peter; Altman, Roger B.; Blanchard, Scott C.; Onuchic, José N.; Sanbonmatsu, Karissa Y. (2010-06-01). "Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways". RNA (New York, N.Y.). 16 (6): 1196–1204. doi:10.1261/rna.2035410. ISSN 1469-9001. PMC 2874171. PMID 20427512.
- ^ Noel, Jeffrey K.; Whitford, Paul C. (2016-10-31). "How EF-Tu can contribute to efficient proofreading of aa-tRNA by the ribosome". Nature Communications. 7: 13314. doi:10.1038/ncomms13314. ISSN 2041-1723. PMC 5095583. PMID 27796304.
- ^ Defeu Soufo, Hervé Joël; Reimold, Christian; Linne, Uwe; Knust, Tobias; Gescher, Johannes; Graumann, Peter L. (2010-02-16). "Bacterial translation elongation factor EF-Tu interacts and colocalizes with actin-like MreB protein". Proceedings of the National Academy of Sciences of the United States of America. 107 (7): 3163–3168. doi:10.1073/pnas.0911979107. ISSN 0027-8424. PMC 2840354. PMID 20133608.
- ^ Mayer, F. (2003-01-01). "Cytoskeletons in prokaryotes". Cell Biology International. 27 (5): 429–438. doi:10.1016/s1065-6995(03)00035-0. ISSN 1065-6995. PMID 12758091. S2CID 40897586.
- ^ Mayer, Frank (2006-01-01). "Cytoskeletal elements in bacteria Mycoplasma pneumoniae, Thermoanaerobacterium sp., and Escherichia coli as revealed by electron microscopy". Journal of Molecular Microbiology and Biotechnology. 11 (3–5): 228–243. doi:10.1159/000094057. ISSN 1464-1801. PMID 16983198. S2CID 23701662.
- ^ Richarme, G. (1998-11-09). "Protein-disulfide isomerase activity of elongation factor EF-Tu". Biochemical and Biophysical Research Communications. 252 (1): 156–161. doi:10.1006/bbrc.1998.9591. ISSN 0006-291X. PMID 9813162.
- ^ Kudlicki, W.; Coffman, A.; Kramer, G.; Hardesty, B. (1997-12-19). "Renaturation of rhodanese by translational elongation factor (EF) Tu. Protein refolding by EF-Tu flexing". The Journal of Biological Chemistry. 272 (51): 32206–32210. doi:10.1074/jbc.272.51.32206. ISSN 0021-9258. PMID 9405422.
- ^ Caldas, T. D.; El Yaagoubi, A.; Kohiyama, M.; Richarme, G. (1998-10-01). "Purification of elongation factors EF-Tu and EF-G from Escherichia coli by covalent chromatography on thiol-sepharose". Protein Expression and Purification. 14 (1): 65–70. doi:10.1006/prep.1998.0922. ISSN 1046-5928. PMID 9758752.
- ^ Wiborg, O.; Andersen, C.; Knudsen, C. R.; Clark, B. F.; Nyborg, J. (1996-08-23). "Mapping Escherichia coli elongation factor Tu residues involved in binding of aminoacyl-tRNA". The Journal of Biological Chemistry. 271 (34): 20406–20411. doi:10.1074/jbc.271.34.20406. ISSN 0021-9258. PMID 8702777.
- ^ Wurmbach, P.; Nierhaus, K. H. (1979-01-01). "Isolation of the protein synthesis elongation factors EF-Tu, EF-Ts, and EF-G from Escherichia coli". Methods in Enzymology. 60: 593–606. doi:10.1016/s0076-6879(79)60056-3. ISSN 0076-6879. PMID 379535.
- ^ Wang, Y.; Jiang, Y.; Meyering-Voss, M.; Sprinzl, M.; Sigler, P. B. (1997-08-01). "Crystal structure of the EF-Tu.EF-Ts complex from Thermus thermophilus". Nature Structural Biology. 4 (8): 650–656. doi:10.1038/nsb0897-650. ISSN 1072-8368. PMID 9253415. S2CID 10644042.
- ^ Nissen, P.; Kjeldgaard, M.; Thirup, S.; Polekhina, G.; Reshetnikova, L.; Clark, B. F.; Nyborg, J. (1995-12-01). "Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog". Science (New York, N.Y.). 270 (5241): 1464–1472. doi:10.1126/science.270.5241.1464. ISSN 0036-8075. PMID 7491491. S2CID 24817616.
- ^ Möller, W.; Schipper, A.; Amons, R. (1987-09-01). "A conserved amino acid sequence around Arg-68 of Artemia elongation factor 1 alpha is involved in the binding of guanine nucleotides and aminoacyl transfer RNAs". Biochimie. 69 (9): 983–989. doi:10.1016/0300-9084(87)90232-x. ISSN 0300-9084. PMID 3126836.
- ^ Kjeldgaard, M.; Nissen, P.; Thirup, S.; Nyborg, J. (1993-09-15). "The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation". Structure (London, England: 1993). 1 (1): 35–50. doi:10.1016/0969-2126(93)90007-4. ISSN 0969-2126. PMID 8069622.
- ^ Selva, E.; Beretta, G.; Montanini, N.; Saddler, G. S.; Gastaldo, L.; Ferrari, P.; Lorenzetti, R.; Landini, P.; Ripamonti, F. (1991-07-01). "Antibiotic GE2270 a: a novel inhibitor of bacterial protein synthesis. I. Isolation and characterization". The Journal of Antibiotics. 44 (7): 693–701. doi:10.7164/antibiotics.44.693. ISSN 0021-8820. PMID 1908853.
- ^ Hogg, T.; Mesters, J. R.; Hilgenfeld, R. (2002-02-01). "Inhibitory mechanisms of antibiotics targeting elongation factor Tu". Current Protein & Peptide Science. 3 (1): 121–131. doi:10.2174/1389203023380855. ISSN 1389-2037. PMID 12370016.
- ^ Andersen, Gregers R.; Nissen, Poul; Nyborg, Jens (2003-08-01). "Elongation factors in protein biosynthesis". Trends in Biochemical Sciences. 28 (8): 434–441. doi:10.1016/S0968-0004(03)00162-2. ISSN 0968-0004. PMID 12932732.
- ^ Parmeggiani, Andrea; Nissen, Poul (2006-08-21). "Elongation factor Tu-targeted antibiotics: four different structures, two mechanisms of action". FEBS Letters. 580 (19): 4576–4581. doi:10.1016/j.febslet.2006.07.039. ISSN 0014-5793. PMID 16876786. S2CID 20811259.
External links[edit]
- Peptide+Elongation+Factor+Tu at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
